Other cell groups


all cells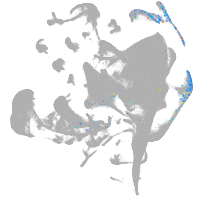 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 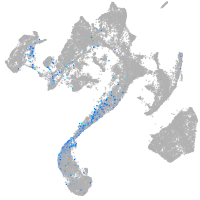 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory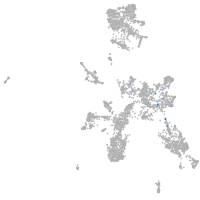 |
otic / lateral line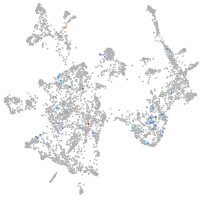 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| txlnbb | 0.250 | hmgb1b | -0.039 |
| rbfox1l | 0.248 | marcksl1a | -0.039 |
| tmx2a | 0.244 | si:ch211-288g17.3 | -0.035 |
| fitm1 | 0.242 | chd4a | -0.031 |
| klhl31 | 0.239 | tuba1c | -0.031 |
| smyd1b | 0.239 | cbx5 | -0.030 |
| txlnba | 0.235 | si:ch211-137a8.4 | -0.030 |
| zgc:101853 | 0.234 | h2afva | -0.029 |
| CABZ01072309.2 | 0.234 | mdkb | -0.029 |
| cacng1a | 0.233 | nova2 | -0.029 |
| klhl40b | 0.233 | pcna | -0.029 |
| desma | 0.231 | stmn1a | -0.029 |
| zgc:92518 | 0.230 | tuba1a | -0.029 |
| tnni2b.1 | 0.229 | ywhaqb | -0.029 |
| CABZ01078594.1 | 0.228 | ctsla | -0.028 |
| srl | 0.228 | anp32a | -0.027 |
| cav3 | 0.227 | gapdhs | -0.027 |
| tgm2a | 0.224 | gpm6aa | -0.027 |
| CABZ01072309.1 | 0.223 | morf4l1 | -0.027 |
| tmem38a | 0.222 | pfn2 | -0.027 |
| klhl41b | 0.221 | rbbp4 | -0.027 |
| si:ch211-131k2.3 | 0.220 | dap1b | -0.027 |
| hsp90aa1.1 | 0.217 | hmgn3 | -0.026 |
| tnnt2c | 0.216 | mcm7 | -0.026 |
| smyd2b | 0.215 | ckbb | -0.025 |
| acta1a | 0.214 | gnb1b | -0.025 |
| actc1a | 0.214 | nutf2l | -0.025 |
| zgc:158296 | 0.212 | seta | -0.025 |
| tnnt3a | 0.211 | si:ch211-156b7.4 | -0.025 |
| chrnd | 0.210 | tmem258 | -0.025 |
| stac3 | 0.210 | zgc:153867 | -0.025 |
| tmem182a | 0.209 | ccni | -0.024 |
| CR376766.2 | 0.208 | dut | -0.024 |
| mybphb | 0.207 | fabp7a | -0.024 |
| aldoab | 0.206 | lbr | -0.024 |




