lysine (K)-specific demethylase 6A
ZFIN 
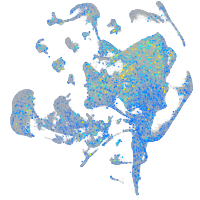
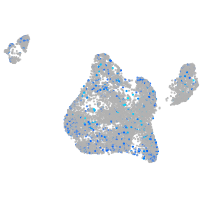
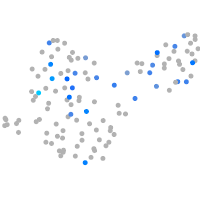
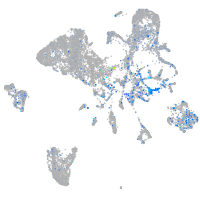
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| top2a | 0.042 | actc1b | -0.021 |
| hmgn2 | 0.041 | mylpfb | -0.021 |
| lbr | 0.040 | cryba1l1 | -0.021 |
| seta | 0.040 | cryba2a | -0.020 |
| cenpf | 0.040 | cryba4 | -0.020 |
| zgc:165555.3 | 0.039 | crybb1 | -0.020 |
| acin1a | 0.039 | si:ch211-214j24.14 | -0.020 |
| mex3b | 0.038 | gnb5b | -0.019 |
| mki67 | 0.038 | crygn2 | -0.019 |
| nucks1a | 0.038 | gpx9 | -0.019 |
| atrx | 0.038 | cryba2b | -0.019 |
| gpr183a | 0.038 | cryba1b | -0.019 |
| kifc1 | 0.038 | pvalb1 | -0.018 |
| tacc3 | 0.038 | crygmx | -0.018 |
| fbxo5 | 0.037 | crybb1l2 | -0.018 |
| cdk1 | 0.037 | crybb1l1 | -0.018 |
| hes6 | 0.037 | lim2.1 | -0.017 |
| baz1b | 0.036 | otc | -0.017 |
| tpx2 | 0.036 | grifin | -0.017 |
| XLOC-038816 | 0.036 | mylpfa | -0.017 |
| foxn4 | 0.036 | crygm2d20 | -0.017 |
| mt-co1 | 0.035 | crygmxl2 | -0.017 |
| baz2ba | 0.035 | crygm2d2 | -0.017 |
| zgc:110216 | 0.035 | mipb | -0.017 |
| ccna2 | 0.035 | prx | -0.017 |
| tuba1a | 0.034 | tnni2a.4 | -0.016 |
| zgc:153409 | 0.034 | crygm2d12 | -0.016 |
| aspm | 0.034 | crygm2d21 | -0.016 |
| tuba8l | 0.034 | pvalb2 | -0.016 |
| celf2 | 0.034 | hsp70.3 | -0.016 |
| si:ch211-288g17.3 | 0.034 | crygm2d10 | -0.016 |
| h3f3a | 0.034 | si:ch1073-303d10.1 | -0.016 |
| anp32a | 0.034 | capn3a | -0.016 |
| smc1al | 0.033 | lim2.4 | -0.016 |
| si:dkey-230p4.1 | 0.033 | rbm24a | -0.016 |