kalirin RhoGEF kinase a
ZFIN 
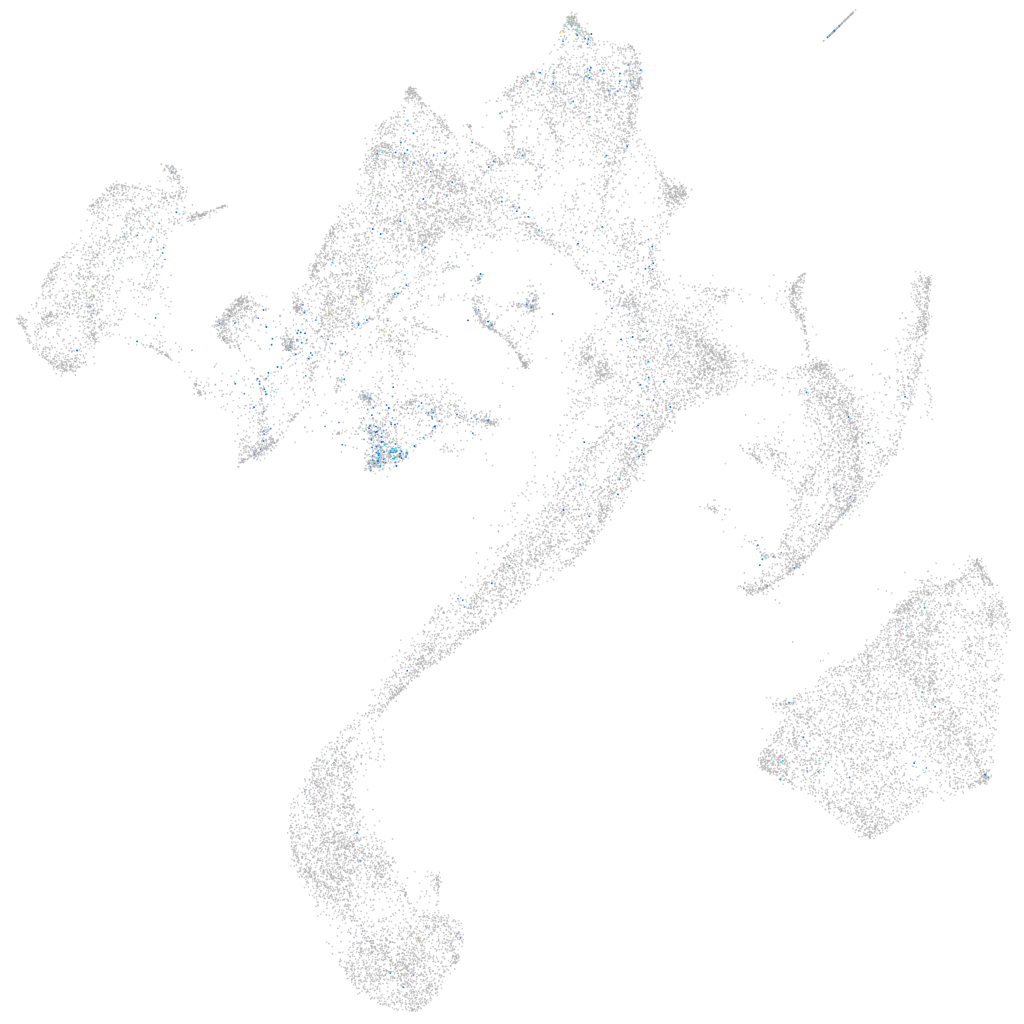
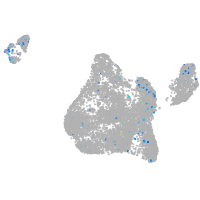
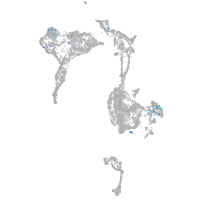
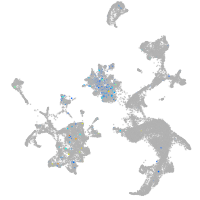
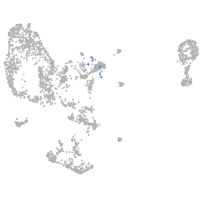
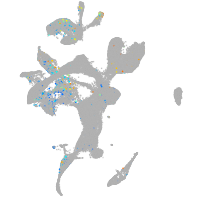
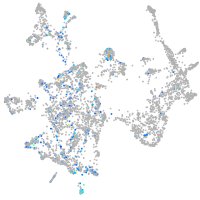
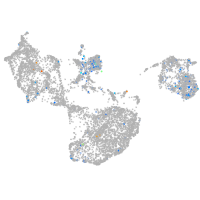
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn1b | 0.189 | tuba8l2 | -0.056 |
| ablim1a | 0.172 | hspb1 | -0.053 |
| fez1 | 0.168 | apoc1 | -0.048 |
| elavl3 | 0.167 | rpl6 | -0.042 |
| gpm6aa | 0.157 | aldoaa | -0.041 |
| tuba1c | 0.150 | apoeb | -0.041 |
| myt1b | 0.141 | eif4ebp3l | -0.041 |
| nova2 | 0.137 | npm1a | -0.040 |
| zgc:65894 | 0.137 | ppp1r3b | -0.040 |
| nsg2 | 0.136 | dkc1 | -0.039 |
| enpp7.1 | 0.130 | cnbpa | -0.038 |
| rtn1b | 0.130 | rsl1d1 | -0.037 |
| gng3 | 0.130 | cdx4 | -0.037 |
| ckbb | 0.129 | plp2 | -0.037 |
| tubb5 | 0.128 | nop2 | -0.036 |
| znf608 | 0.128 | BX927258.1 | -0.036 |
| lhx9 | 0.125 | egfl6 | -0.035 |
| PRKAR2B | 0.125 | tbx16 | -0.035 |
| aplp1 | 0.125 | bxdc2 | -0.035 |
| bcl11ba | 0.125 | ubap2b | -0.035 |
| prrt1 | 0.124 | si:ch211-217k17.7 | -0.034 |
| scrt2 | 0.123 | mrto4 | -0.034 |
| elavl4 | 0.123 | msgn1 | -0.034 |
| stxbp1a | 0.123 | ved | -0.034 |
| thsd7aa | 0.122 | gar1 | -0.034 |
| jagn1a | 0.122 | ebna1bp2 | -0.034 |
| gap43 | 0.120 | cdx1a | -0.033 |
| sept3 | 0.118 | pprc1 | -0.033 |
| myt1la | 0.117 | BX001014.2 | -0.033 |
| vamp2 | 0.116 | cycsb | -0.033 |
| csdc2a | 0.115 | hoxb7a | -0.033 |
| islr2 | 0.114 | polr3gla | -0.033 |
| CT027772.2 | 0.113 | nop58 | -0.033 |
| ppp1r14ba | 0.113 | wu:fb97g03 | -0.033 |
| ptmaa | 0.112 | NC-002333.4 | -0.033 |