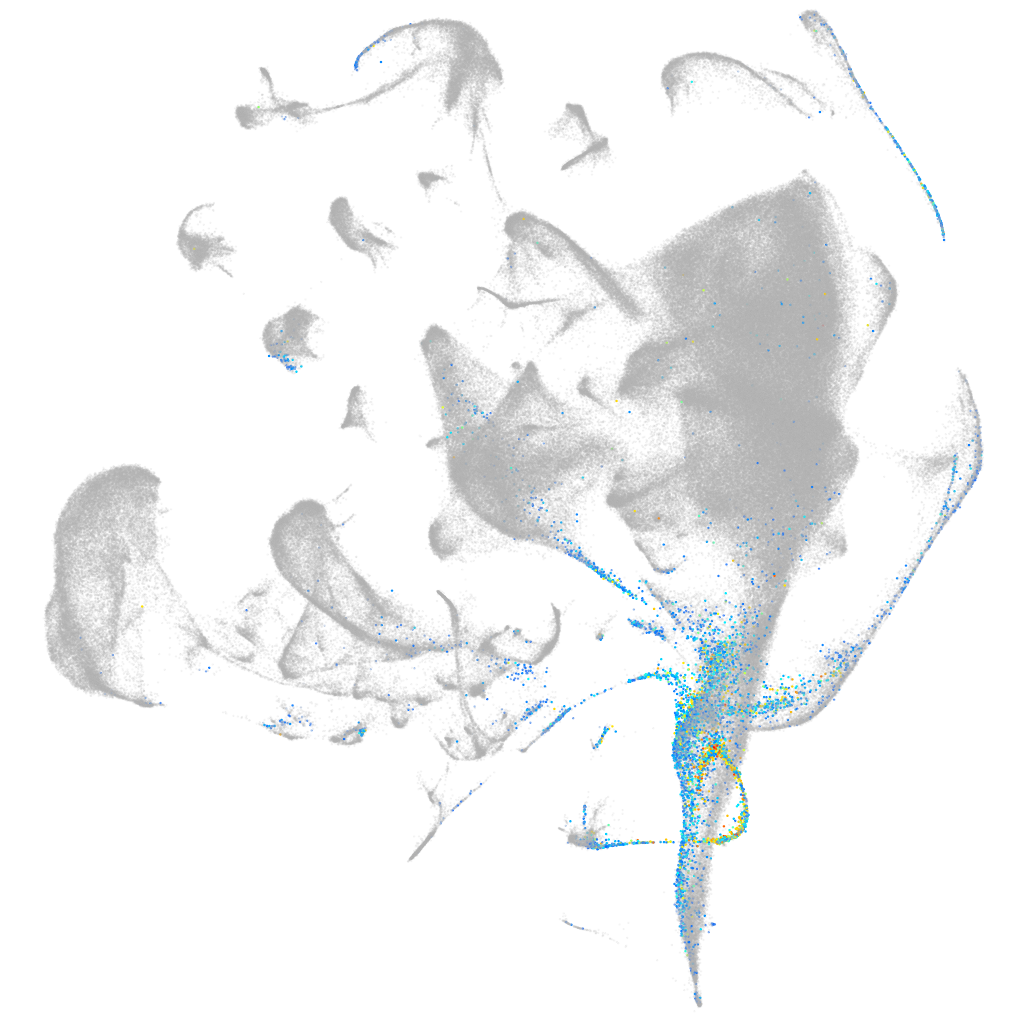
preproinsulin b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm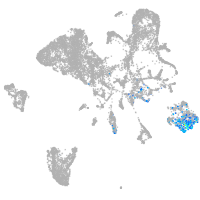 |
axial mesoderm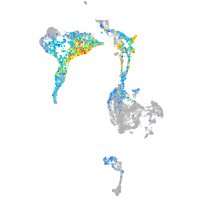 |
muscle 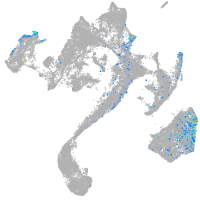 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 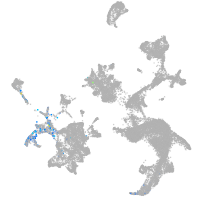 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis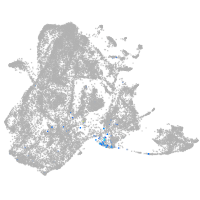 |
periderm |
fin |
ionocytes / mucous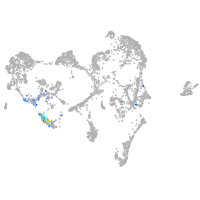 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxa | 0.304 | tuba1c | -0.093 |
| tbxta | 0.282 | gpm6aa | -0.085 |
| ndr2 | 0.253 | gapdhs | -0.081 |
| si:dkeyp-2e4.3 | 0.209 | pvalb1 | -0.079 |
| wu:fb97g03 | 0.209 | fabp3 | -0.078 |
| lft1 | 0.208 | prdx2 | -0.078 |
| hspb1 | 0.207 | rtn1a | -0.078 |
| ntd5 | 0.204 | hbbe1.3 | -0.077 |
| fadd | 0.201 | nova2 | -0.077 |
| chrd | 0.200 | stmn1b | -0.077 |
| XLOC-041707 | 0.187 | calm1a | -0.076 |
| loxl5b | 0.186 | ckbb | -0.075 |
| asph | 0.185 | pvalb2 | -0.075 |
| noto | 0.183 | rnasekb | -0.075 |
| XLOC-002640 | 0.182 | ccni | -0.074 |
| lft2 | 0.180 | atp6v0cb | -0.073 |
| tbx16 | 0.179 | elavl3 | -0.073 |
| tbx3b | 0.178 | hbae3 | -0.073 |
| admp | 0.176 | mdkb | -0.073 |
| nog1 | 0.176 | krt4 | -0.071 |
| ripply1 | 0.176 | marcksl1a | -0.069 |
| frzb | 0.171 | cyt1 | -0.068 |
| rrbp1b | 0.171 | gng3 | -0.068 |
| foxa3 | 0.170 | hbae1.1 | -0.068 |
| aplnra | 0.169 | sncb | -0.067 |
| wnt11f2 | 0.169 | gpm6ab | -0.065 |
| akap12b | 0.166 | vamp2 | -0.065 |
| p4ha2 | 0.166 | cspg5a | -0.062 |
| foxa2 | 0.165 | cyt1l | -0.062 |
| col15a1a | 0.163 | si:ch1073-429i10.3.1 | -0.062 |
| stm | 0.163 | hbbe1.1 | -0.061 |
| plod1a | 0.162 | elavl4 | -0.059 |
| plod2 | 0.161 | stx1b | -0.059 |
| emilin3a | 0.160 | tmsb | -0.059 |
| lrrc59 | 0.160 | celf2 | -0.058 |