preproinsulin b
ZFIN 
Other cell groups


all cells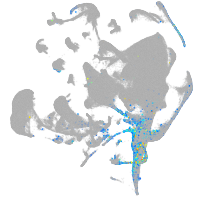 |
blastomeres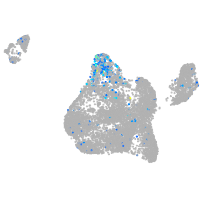 |
PGCs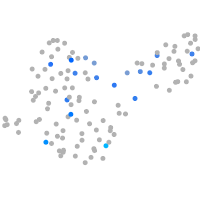 |
endoderm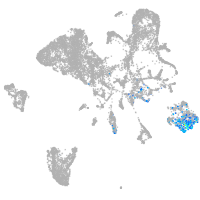 |
axial mesoderm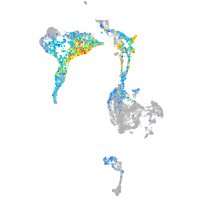 |
muscle 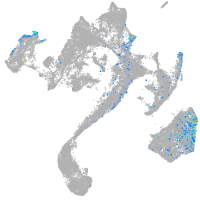 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 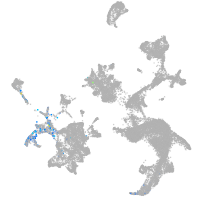 |
pronephros 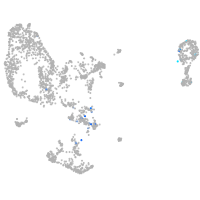 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| twist2 | 0.347 | fabp3 | -0.067 |
| rbp7b | 0.278 | tuba1c | -0.046 |
| ptx3a | 0.268 | nova2 | -0.042 |
| aplnr2 | 0.234 | gpm6aa | -0.041 |
| ntd5 | 0.225 | nme2b.1 | -0.041 |
| fgfrl1b | 0.225 | mdkb | -0.040 |
| plod2 | 0.221 | ptmaa | -0.036 |
| col15a1a | 0.213 | si:dkey-56m19.5 | -0.035 |
| spon2a | 0.212 | fabp7a | -0.034 |
| AL954327.1 | 0.208 | tmsb4x | -0.032 |
| col8a1a | 0.203 | sox3 | -0.031 |
| tmem119a | 0.199 | prdx2 | -0.031 |
| slc6a4a | 0.198 | elavl3 | -0.031 |
| pappaa | 0.198 | vim | -0.031 |
| mfrp | 0.193 | tuba1a | -0.031 |
| hapln1b | 0.191 | CU467822.1 | -0.030 |
| angpt1 | 0.190 | actc1b | -0.030 |
| optc | 0.189 | pvalb2 | -0.030 |
| CT033842.1 | 0.183 | rtn1a | -0.030 |
| col9a1b | 0.181 | zgc:165461 | -0.029 |
| kng1 | 0.177 | atp1a1b | -0.029 |
| ism2a | 0.175 | CU634008.1 | -0.029 |
| lamb1a | 0.174 | ckbb | -0.029 |
| si:ch211-157b11.8 | 0.173 | gpm6bb | -0.028 |
| fbn2b | 0.170 | hmgb3a | -0.028 |
| rcn3 | 0.167 | si:ch73-21g5.7 | -0.028 |
| fkbp7 | 0.166 | COX3 | -0.028 |
| apela | 0.163 | pvalb1 | -0.027 |
| rrbp1b | 0.161 | hmgb1a | -0.027 |
| gdf6b | 0.161 | hmgb1b | -0.026 |
| timp4.1 | 0.160 | lfng | -0.026 |
| serpinh1b | 0.158 | cspg5a | -0.026 |
| kazald2 | 0.156 | rplp2 | -0.026 |
| frem3 | 0.155 | hbbe1.3 | -0.025 |
| si:ch73-361h17.1 | 0.154 | tmsb | -0.025 |




