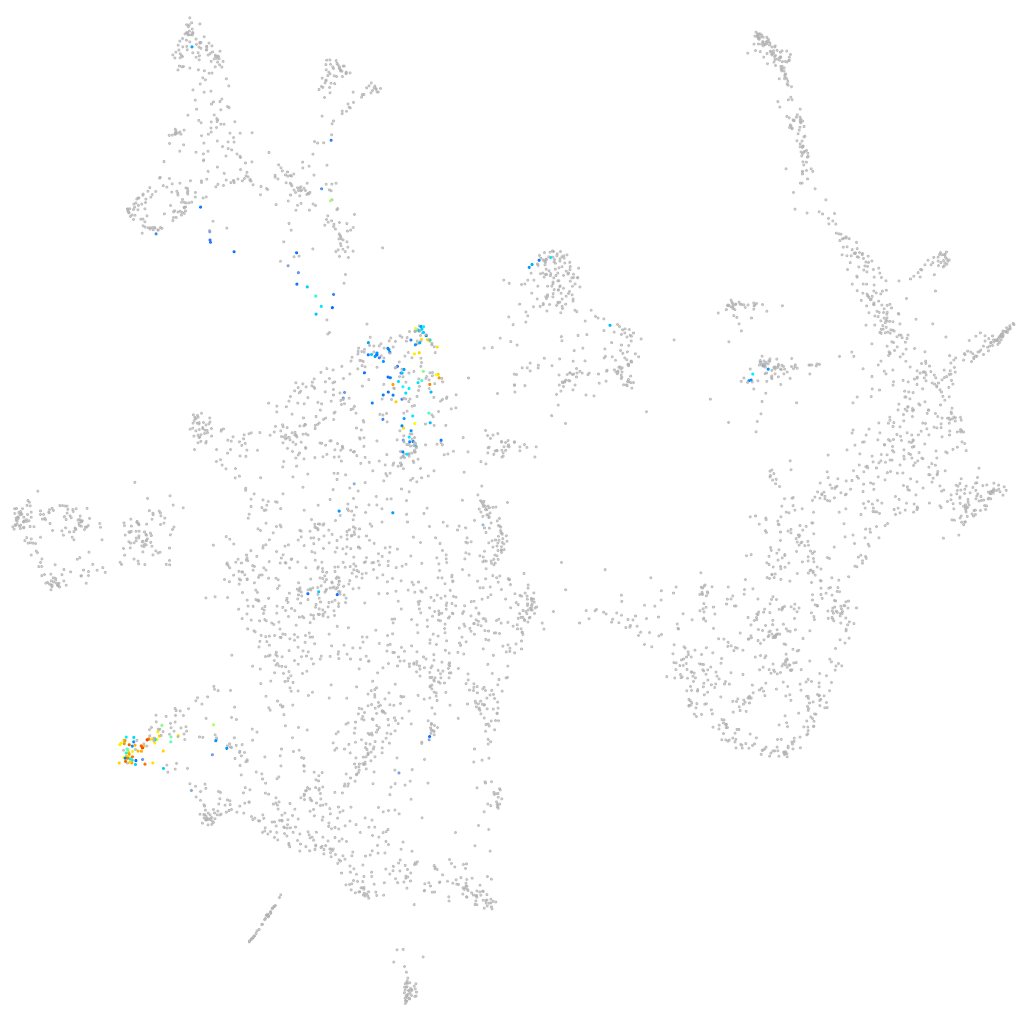
"HERV-H LTR-associating 1, transcript variant X6"
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line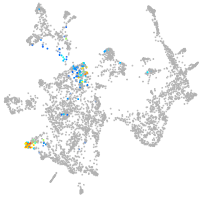 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zpld1b | 0.617 | krt8 | -0.137 |
| zpld1a | 0.495 | dynll1 | -0.122 |
| kcnq5a | 0.475 | krt18a.1 | -0.105 |
| glula | 0.454 | fkbp1aa | -0.104 |
| msx3 | 0.426 | snrpd1 | -0.104 |
| slc13a5a | 0.423 | gsnb | -0.100 |
| slc25a22b | 0.422 | npm1a | -0.099 |
| zwi | 0.399 | marcksl1b | -0.099 |
| mb | 0.385 | cldnh | -0.097 |
| cx30.3 | 0.381 | tubb2b | -0.096 |
| cyp26b1 | 0.370 | krt15 | -0.095 |
| otog | 0.367 | abracl | -0.094 |
| isl1 | 0.348 | dlx3b | -0.092 |
| FAM163A | 0.347 | gpc1a | -0.092 |
| si:ch211-236l14.4 | 0.340 | krt18b | -0.091 |
| igfbp3 | 0.330 | snrpe | -0.091 |
| cryabb | 0.327 | wu:fb18f06 | -0.088 |
| adcy2b | 0.323 | nqo1 | -0.088 |
| nfasca | 0.321 | cx43.4 | -0.087 |
| dusp14 | 0.320 | tuba8l4 | -0.087 |
| mia | 0.318 | si:ch211-222l21.1 | -0.087 |
| aif1l | 0.310 | tfap2a | -0.087 |
| tmem47 | 0.307 | capn8 | -0.087 |
| tectb | 0.306 | six2b | -0.085 |
| myo18aa | 0.305 | ilf2 | -0.084 |
| trim101 | 0.303 | ssrp1a | -0.084 |
| slc1a3a | 0.296 | stmn1a | -0.084 |
| kitlga | 0.293 | si:ch211-195b11.3 | -0.082 |
| mdkb | 0.292 | txn | -0.082 |
| efhd1 | 0.291 | ahnak | -0.081 |
| fads2 | 0.284 | hmga1a | -0.081 |
| FO704813.1 | 0.283 | mcama | -0.080 |
| dlx1a | 0.282 | krt4 | -0.080 |
| slc16a1a | 0.281 | znf385a | -0.080 |
| adgrg3 | 0.280 | si:ch211-39i22.1 | -0.079 |