G protein-coupled receptor kinase 7a
ZFIN 
Other cell groups


all cells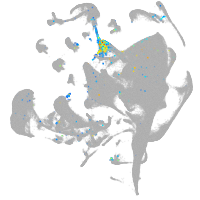 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 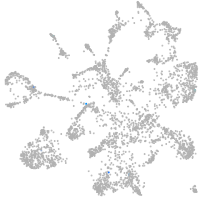 |
mesenchyme 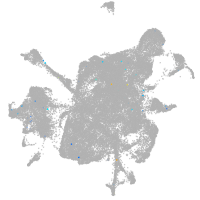 |
hematopoietic / vasculature 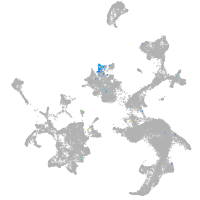 |
pronephros 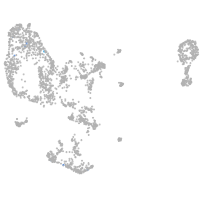 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm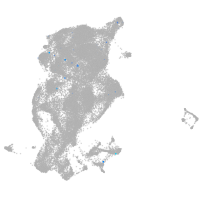 |
fin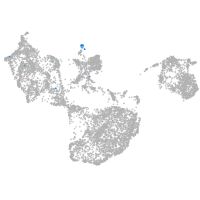 |
ionocytes / mucous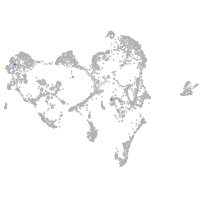 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc1a2a | 0.319 | serbp1a | -0.025 |
| zgc:193593 | 0.306 | cnbpa | -0.023 |
| gnat2 | 0.294 | ube2v2 | -0.022 |
| si:ch1073-469d17.2 | 0.289 | hnrnpa0a | -0.021 |
| prph2a | 0.284 | dkc1 | -0.020 |
| impg1a | 0.277 | pfdn4 | -0.019 |
| opn1lw2 | 0.276 | pprc1 | -0.019 |
| prph2b | 0.276 | ddx39ab | -0.019 |
| guca1c | 0.273 | fbl | -0.019 |
| camk1ga | 0.270 | dnajc1 | -0.019 |
| cnga3a | 0.266 | hdgfl2 | -0.019 |
| CT573282.1 | 0.259 | mak16 | -0.019 |
| gnb3b | 0.258 | rsl1d1 | -0.018 |
| rcvrn3 | 0.254 | mrto4 | -0.018 |
| arr3a | 0.251 | syncrip | -0.018 |
| rcvrn2 | 0.247 | aldoaa | -0.018 |
| si:dkey-1c7.1 | 0.245 | eif2s2 | -0.018 |
| pde6ha | 0.242 | eif3ha | -0.017 |
| pde6c | 0.239 | ptges3b | -0.017 |
| prph2l | 0.238 | srsf2a | -0.017 |
| si:dkey-17e16.15 | 0.237 | gar1 | -0.017 |
| cnga3b | 0.235 | hmga1a | -0.017 |
| rbp4l | 0.234 | rwdd1 | -0.017 |
| SH3TC1 | 0.232 | pa2g4a | -0.017 |
| zgc:73359 | 0.231 | asph | -0.017 |
| grk1b | 0.230 | rcl1 | -0.017 |
| guk1b | 0.227 | snrpd2 | -0.017 |
| guca1g | 0.222 | bxdc2 | -0.017 |
| impg2a | 0.221 | srsf5a | -0.016 |
| pdcb | 0.216 | adi1 | -0.016 |
| FO818704.1 | 0.210 | taf15 | -0.016 |
| zgc:162331 | 0.204 | ppan | -0.016 |
| si:ch211-207l14.1 | 0.203 | eif4g1a | -0.016 |
| FO904901.1 | 0.198 | riox2 | -0.016 |
| rcvrna | 0.197 | ppp2cb | -0.016 |




