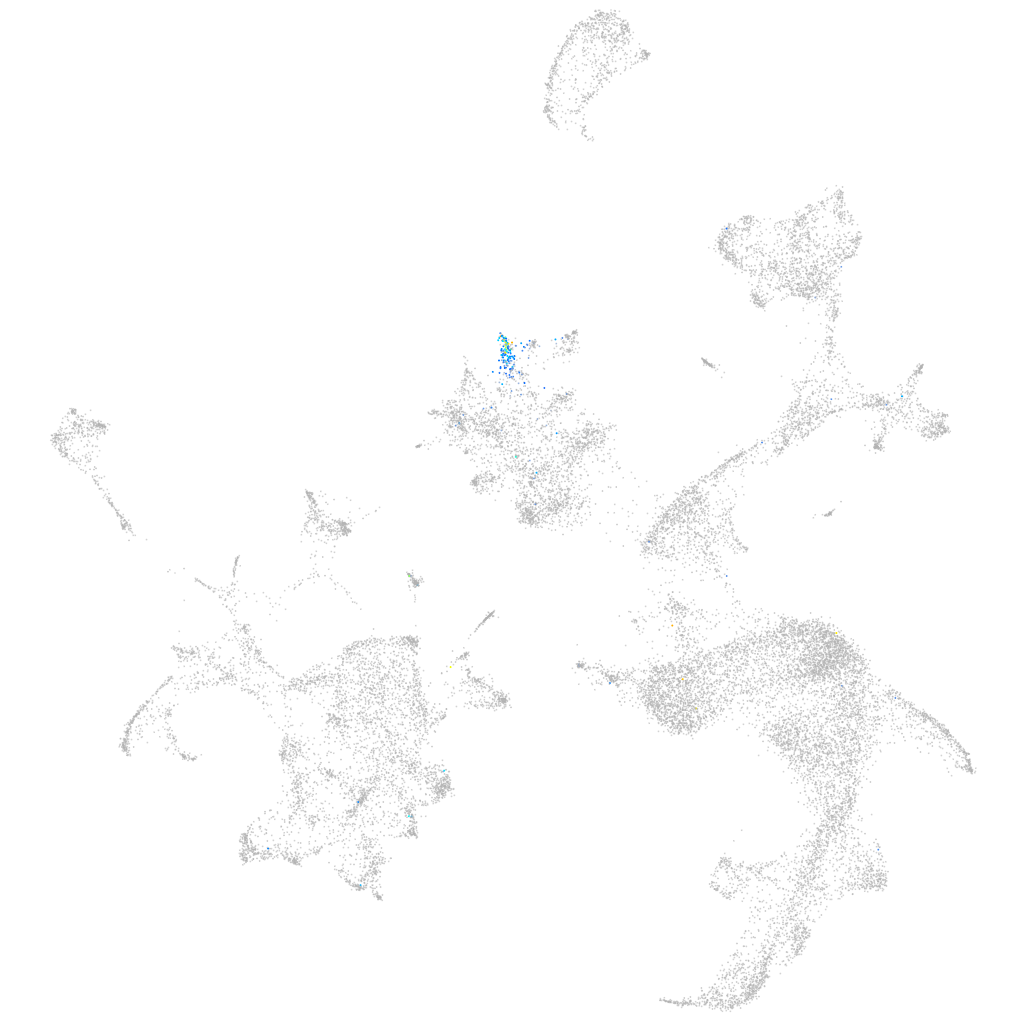
G protein-coupled receptor kinase 7a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 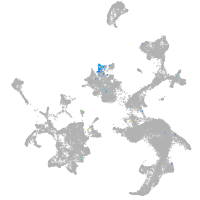 |
pronephros 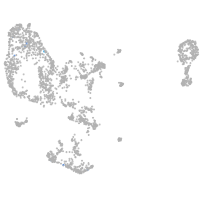 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm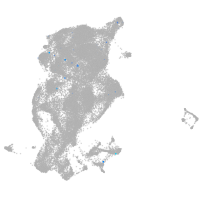 |
fin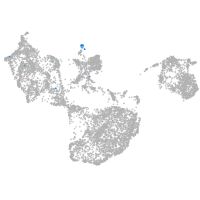 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prph2a | 0.585 | hbbe3 | -0.038 |
| cngb3.2 | 0.583 | zgc:56493 | -0.035 |
| rcvrn2 | 0.582 | rpl9 | -0.032 |
| pdcb | 0.581 | arhgdia | -0.031 |
| gnat2 | 0.574 | pfn1 | -0.031 |
| rcvrn3 | 0.571 | lmo2 | -0.029 |
| grk1b | 0.559 | arpc1b | -0.026 |
| zgc:73359 | 0.559 | bzw1b | -0.026 |
| cnga3a | 0.554 | mob1a | -0.026 |
| gnb3b | 0.548 | rnaseka | -0.026 |
| guk1b | 0.544 | si:dkey-112e7.2 | -0.026 |
| ckmt2a | 0.543 | msna | -0.026 |
| arr3a | 0.540 | anp32b | -0.025 |
| pde6c | 0.540 | hhex | -0.025 |
| si:ch1073-469d17.2 | 0.536 | arl4aa | -0.025 |
| slc25a3a | 0.535 | rps12 | -0.025 |
| si:ch211-81a5.8 | 0.529 | snx1a | -0.025 |
| si:ch211-285j22.3 | 0.525 | rplp0 | -0.025 |
| si:dkey-17e16.15 | 0.520 | laptm5 | -0.025 |
| guca1c | 0.517 | rhocb | -0.024 |
| rgs9a | 0.516 | coro1a | -0.024 |
| rbp4l | 0.508 | etf1b | -0.024 |
| arl3l2 | 0.501 | CT030188.1 | -0.024 |
| pde6h | 0.493 | snx5 | -0.024 |
| tmem237a | 0.491 | rac2 | -0.024 |
| cnga3b | 0.491 | urod | -0.024 |
| prph2b | 0.486 | tpm3 | -0.024 |
| rcvrna | 0.467 | nrros | -0.024 |
| si:ch1073-303d10.1 | 0.467 | capgb | -0.023 |
| arl3l1 | 0.464 | spi1b | -0.023 |
| tmx3a | 0.460 | cst14a.2 | -0.023 |
| atp1b2b | 0.458 | srgn | -0.023 |
| syt5a | 0.456 | she | -0.023 |
| gucy2d | 0.451 | yrk | -0.022 |
| gabra6a | 0.449 | mafbb | -0.022 |