G protein-coupled receptor kinase 7a
ZFIN 
Other cell groups


all cells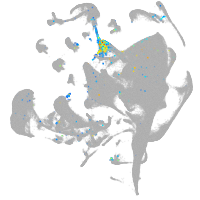 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 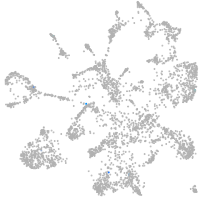 |
mesenchyme 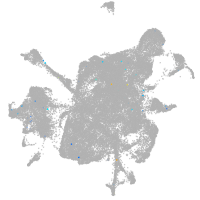 |
hematopoietic / vasculature 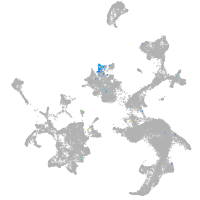 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prph2a | 0.767 | rtn1a | -0.063 |
| gnat2 | 0.764 | gnb1b | -0.060 |
| pdcb | 0.760 | marcksb | -0.060 |
| gnb3b | 0.751 | calm2a | -0.057 |
| pde6c | 0.744 | rtn1b | -0.057 |
| zgc:73359 | 0.732 | si:ch211-288g17.3 | -0.056 |
| grk1b | 0.726 | tmeff1b | -0.056 |
| rgs9a | 0.713 | nova2 | -0.055 |
| slc25a3a | 0.706 | si:dkey-56m19.5 | -0.055 |
| cnga3a | 0.688 | si:ch73-46j18.5 | -0.054 |
| rcvrn3 | 0.688 | tubb5 | -0.054 |
| prph2b | 0.684 | cd99l2 | -0.053 |
| guk1b | 0.682 | ctsla | -0.053 |
| si:ch211-81a5.8 | 0.667 | nucks1a | -0.053 |
| si:dkey-17e16.15 | 0.666 | zc4h2 | -0.053 |
| guca1c | 0.654 | tmem59l | -0.051 |
| rcvrn2 | 0.653 | fam168a | -0.049 |
| ckmt2a | 0.645 | gpm6aa | -0.049 |
| si:ch1073-469d17.2 | 0.645 | ywhag2 | -0.049 |
| cngb3.2 | 0.644 | atp2b1a | -0.048 |
| gngt2b | 0.635 | ctnnb1 | -0.048 |
| si:ch211-285j22.3 | 0.635 | elavl4 | -0.048 |
| si:ch1073-303d10.1 | 0.634 | gng3 | -0.048 |
| si:ch211-207l14.1 | 0.634 | nsg2 | -0.048 |
| si:cabz01076231.1 | 0.618 | stx1b | -0.048 |
| arr3a | 0.614 | atp2b2 | -0.047 |
| tmem237a | 0.613 | elavl3 | -0.047 |
| arl3l2 | 0.605 | hnrnpa1a | -0.047 |
| impg2a | 0.600 | stmn2a | -0.047 |
| slc24a2 | 0.596 | sv2a | -0.047 |
| cnga3b | 0.596 | marcksl1a | -0.047 |
| syt5a | 0.585 | chd4a | -0.046 |
| rbp4l | 0.584 | syt2a | -0.046 |
| sv2ba | 0.569 | ywhaz | -0.046 |
| si:rp71-39b20.4 | 0.558 | stxbp1a | -0.046 |




