Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin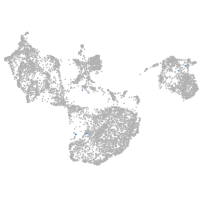 |
ionocytes / mucous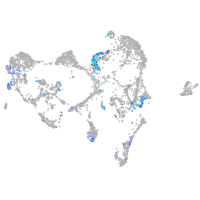 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dab2 | 0.248 | tuba1c | -0.067 |
| si:dkey-28n18.9 | 0.246 | hmgb1b | -0.063 |
| amn | 0.225 | gpm6aa | -0.062 |
| iqgap2 | 0.198 | mdkb | -0.058 |
| si:dkey-194e6.1 | 0.198 | tuba1a | -0.056 |
| mrc1a | 0.197 | rtn1a | -0.054 |
| LOC101885014 | 0.195 | hmgb3a | -0.053 |
| dpys | 0.193 | atp6v0cb | -0.052 |
| dpydb | 0.191 | elavl3 | -0.052 |
| cx32.3 | 0.190 | marcksb | -0.052 |
| acmsd | 0.188 | tmeff1b | -0.052 |
| haao | 0.187 | ckbb | -0.051 |
| kynu | 0.187 | si:ch211-137a8.4 | -0.051 |
| si:ch211-117n7.6 | 0.186 | stmn1b | -0.049 |
| slc2a2 | 0.183 | tubb5 | -0.049 |
| aspdh | 0.182 | gpm6ab | -0.048 |
| TMEM236 | 0.182 | nova2 | -0.048 |
| lyve1b | 0.181 | cspg5a | -0.047 |
| hao2 | 0.180 | cadm3 | -0.046 |
| gpr182 | 0.179 | celf2 | -0.046 |
| ftcd | 0.177 | si:dkey-276j7.1 | -0.045 |
| clic5b | 0.176 | zc4h2 | -0.045 |
| g6pca.2 | 0.175 | mdka | -0.044 |
| grhprb | 0.175 | rnasekb | -0.044 |
| si:dkey-197j19.5 | 0.174 | epb41a | -0.043 |
| zgc:194392 | 0.174 | fam168a | -0.043 |
| etv2 | 0.173 | hnrnpa0a | -0.043 |
| pbld | 0.173 | rtn1b | -0.043 |
| stab2 | 0.173 | si:ch211-133n4.4 | -0.043 |
| fbp1b | 0.172 | sncb | -0.043 |
| si:ch73-185c24.2 | 0.172 | atpv0e2 | -0.041 |
| st3gal7 | 0.172 | nat8l | -0.041 |
| tmprss2 | 0.172 | si:ch211-288g17.3 | -0.041 |
| ezra | 0.171 | atp1b2a | -0.040 |
| agxta | 0.170 | fabp7a | -0.040 |




