hydroxyacid oxidase 2 (long chain)
ZFIN 
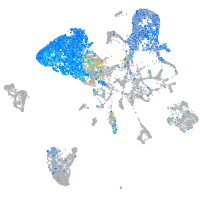
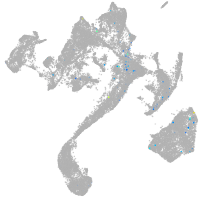
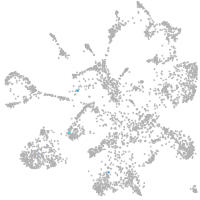
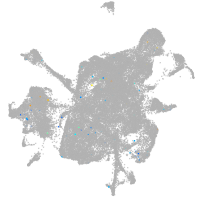
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grhprb | 0.637 | marcksl1a | -0.102 |
| hao1 | 0.637 | tuba1c | -0.097 |
| ftcd | 0.623 | hmgb1b | -0.093 |
| agxta | 0.621 | nova2 | -0.091 |
| fbp1b | 0.617 | gpm6aa | -0.090 |
| acmsd | 0.611 | hmgb3a | -0.089 |
| g6pca.2 | 0.611 | jpt1b | -0.087 |
| dpys | 0.606 | atp6v0cb | -0.085 |
| cx32.3 | 0.602 | gpm6ab | -0.084 |
| f2 | 0.597 | rtn1a | -0.083 |
| si:dkey-197j19.5 | 0.597 | hnrnpa0a | -0.081 |
| dpydb | 0.591 | tubb5 | -0.080 |
| slc2a2 | 0.590 | celf2 | -0.079 |
| agxtb | 0.589 | ckbb | -0.079 |
| tymp | 0.583 | gapdhs | -0.079 |
| haao | 0.581 | rnasekb | -0.079 |
| pklr | 0.581 | mdkb | -0.077 |
| serpinf2b | 0.580 | tmeff1b | -0.077 |
| rdh1 | 0.575 | tuba1a | -0.077 |
| etnppl | 0.574 | elavl3 | -0.076 |
| serpinf2a | 0.568 | fam168a | -0.075 |
| hal | 0.566 | marcksb | -0.075 |
| pbld | 0.564 | stmn1b | -0.073 |
| si:ch211-288g17.4 | 0.563 | CABZ01080702.1 | -0.071 |
| tdo2a | 0.562 | si:dkey-276j7.1 | -0.070 |
| kng1 | 0.560 | vdac1 | -0.070 |
| aspdh | 0.559 | si:ch1073-429i10.3.1 | -0.070 |
| c9 | 0.559 | chd4a | -0.069 |
| cfb | 0.559 | zc4h2 | -0.069 |
| shbg | 0.558 | atrx | -0.068 |
| f7 | 0.558 | rtn1b | -0.068 |
| vtna | 0.557 | cspg5a | -0.067 |
| zgc:112265 | 0.556 | sncb | -0.067 |
| cyp4v8 | 0.555 | cadm3 | -0.066 |
| plg | 0.555 | si:ch211-137a8.4 | -0.066 |