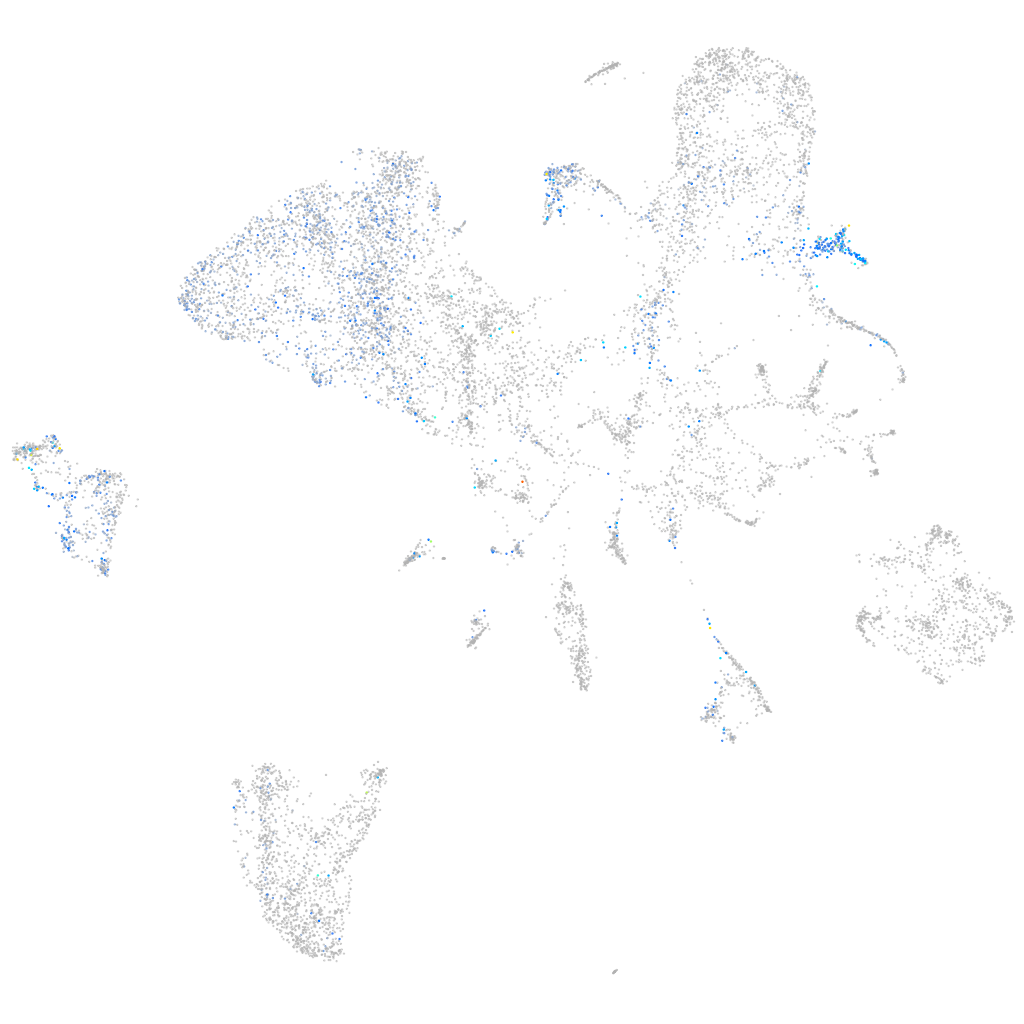
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line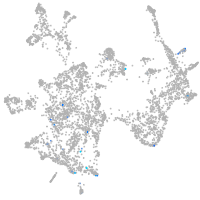 |
epidermis |
periderm |
fin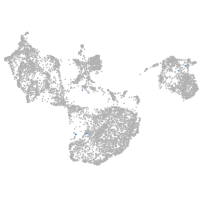 |
ionocytes / mucous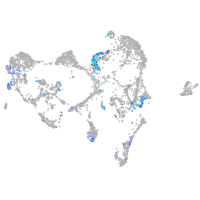 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrp2b | 0.321 | marcksl1b | -0.107 |
| LOC100537272 | 0.314 | rtn1a | -0.106 |
| cubn | 0.312 | hspb1 | -0.098 |
| ctsbb | 0.302 | fabp1b.1 | -0.097 |
| dab2 | 0.299 | akap12b | -0.093 |
| amn | 0.291 | marcksb | -0.092 |
| xirp1 | 0.283 | si:ch211-152c2.3 | -0.086 |
| sptbn5 | 0.269 | fep15 | -0.083 |
| tfeb | 0.258 | gata5 | -0.083 |
| slc6a18 | 0.248 | cdh6 | -0.083 |
| naga | 0.242 | zgc:112160 | -0.083 |
| NAT16 (1 of many) | 0.236 | cx43.4 | -0.083 |
| lgmn | 0.224 | sycn.2 | -0.083 |
| scpep1 | 0.223 | prss59.1 | -0.082 |
| ctsl.1 | 0.222 | CELA1 (1 of many) | -0.082 |
| snx2 | 0.216 | cel.1 | -0.082 |
| slc15a2 | 0.208 | cpa5 | -0.082 |
| tmigd1 | 0.198 | ctrb1 | -0.082 |
| rab32b | 0.198 | apoda.2 | -0.082 |
| si:dkey-28n18.9 | 0.197 | erp27 | -0.081 |
| ctsz | 0.197 | prss59.2 | -0.081 |
| si:ch211-262i1.3 | 0.197 | cpb1 | -0.081 |
| CU499336.2 | 0.196 | ela2 | -0.081 |
| cdaa | 0.196 | ctrl | -0.081 |
| ddit3 | 0.194 | si:ch211-240l19.5 | -0.080 |
| si:dkey-194e6.1 | 0.193 | pdia2 | -0.080 |
| si:ch211-122f10.4 | 0.191 | aif1l | -0.080 |
| fabp6 | 0.190 | cel.2 | -0.080 |
| dpep1 | 0.188 | ela2l | -0.079 |
| egr1 | 0.187 | apoeb | -0.079 |
| agr2 | 0.187 | prss1 | -0.079 |
| gnsb | 0.184 | zgc:136461 | -0.078 |
| CR381686.5 | 0.183 | cpa4 | -0.078 |
| tspan34 | 0.182 | ela3l | -0.077 |
| mfsd12a | 0.182 | cd81a | -0.077 |