glycerol kinase 5
ZFIN 
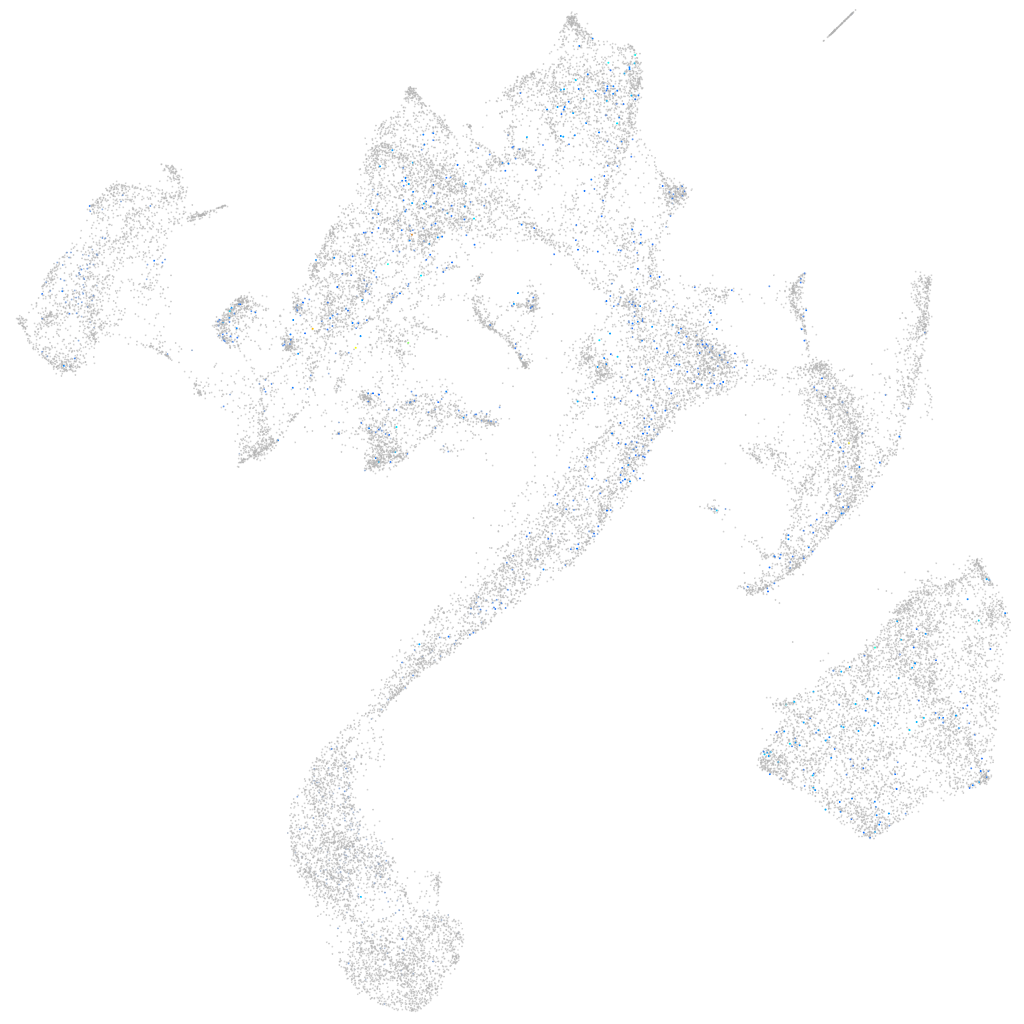
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101882555 | 0.092 | tnnc2 | -0.043 |
| LOC103909845 | 0.077 | tpma | -0.042 |
| ptk7b | 0.076 | actc1b | -0.040 |
| LOC108190036 | 0.072 | acta1b | -0.040 |
| XLOC-034584 | 0.071 | neb | -0.039 |
| pcna | 0.068 | ak1 | -0.037 |
| stmn1a | 0.066 | mylz3 | -0.037 |
| rrm1 | 0.064 | actn3b | -0.037 |
| dut | 0.064 | mybphb | -0.037 |
| elnb | 0.062 | mylpfa | -0.037 |
| tubb2b | 0.061 | gapdh | -0.037 |
| calm3b | 0.060 | ldb3b | -0.036 |
| ranbp1 | 0.060 | acta1a | -0.035 |
| h2afva | 0.058 | smyd1a | -0.035 |
| slc5a9 | 0.057 | pvalb2 | -0.035 |
| LOC108192086 | 0.057 | ckmb | -0.035 |
| kif16bb | 0.057 | hhatla | -0.035 |
| si:ch211-156b7.4 | 0.057 | mylpfb | -0.035 |
| tuba8l4 | 0.057 | ldb3a | -0.035 |
| LOC110439453 | 0.056 | tmod4 | -0.035 |
| cfl1 | 0.056 | myhz2 | -0.035 |
| si:dkey-12f6.5 | 0.055 | tnni2a.4 | -0.035 |
| snrpg | 0.055 | xirp2a | -0.035 |
| prim2 | 0.054 | pvalb1 | -0.034 |
| degs2 | 0.054 | eef2l2 | -0.034 |
| sub1a | 0.054 | tnnt3b | -0.034 |
| sumo3b | 0.054 | rtn2a | -0.034 |
| rbbp4 | 0.054 | klhl43 | -0.034 |
| nutf2l | 0.053 | myom1a | -0.034 |
| tuba8l | 0.053 | ampd1 | -0.034 |
| pa2g4b | 0.053 | prr33 | -0.034 |
| dnajc9 | 0.052 | myom2a | -0.033 |
| rrm2 | 0.052 | ckma | -0.033 |
| pfn2 | 0.052 | si:ch73-367p23.2 | -0.033 |
| pfn2l | 0.052 | klhl31 | -0.033 |