FH2 domain containing 2
ZFIN 
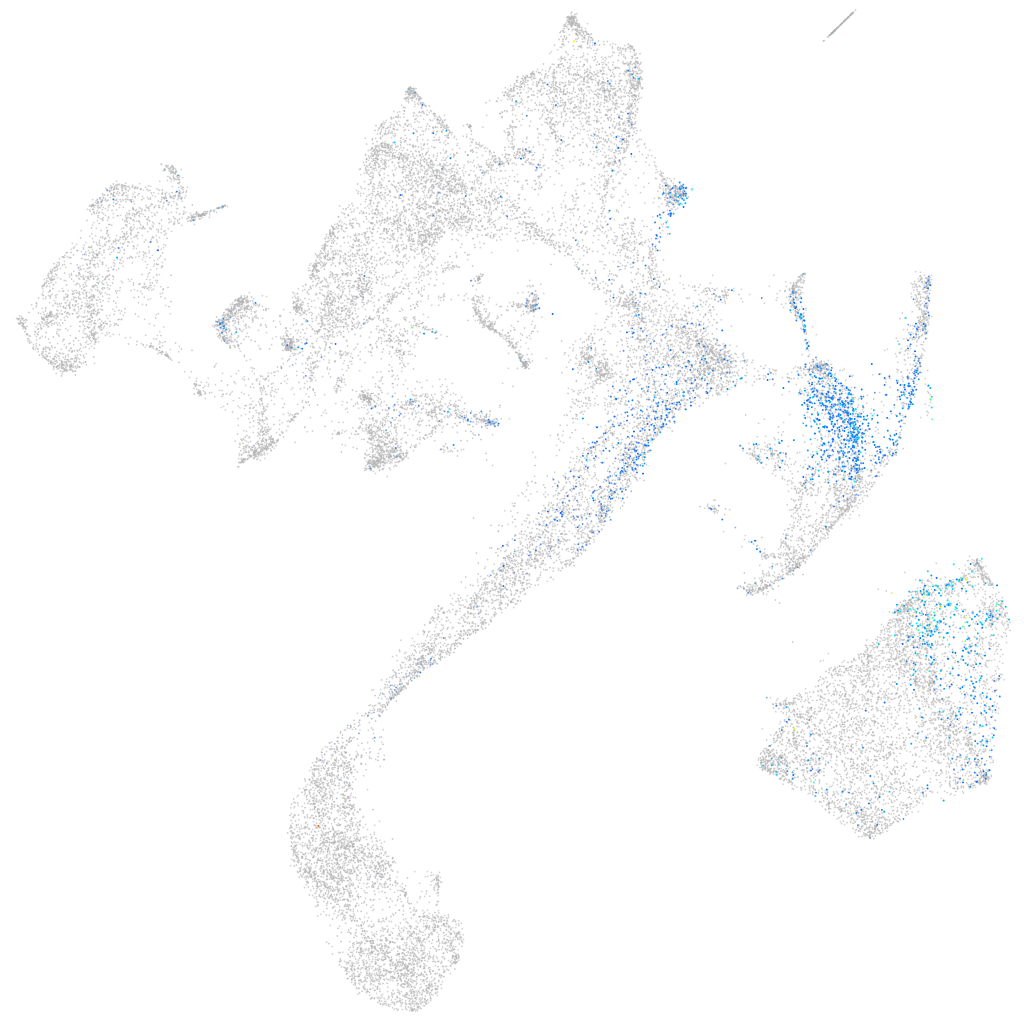
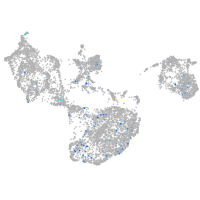
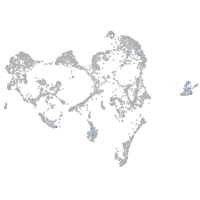
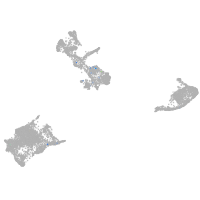
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tbx6 | 0.369 | fabp3 | -0.163 |
| myf5 | 0.264 | bhmt | -0.149 |
| msgn1 | 0.236 | actc1b | -0.146 |
| her1 | 0.234 | ckma | -0.139 |
| grin2da | 0.229 | ckmb | -0.137 |
| gnaia | 0.229 | ak1 | -0.135 |
| apoc1 | 0.227 | atp2a1 | -0.134 |
| fn1b | 0.226 | eef1da | -0.133 |
| pcdh8 | 0.225 | mylpfa | -0.131 |
| efnb2b | 0.225 | pabpc4 | -0.128 |
| ripply2 | 0.225 | si:dkey-16p21.8 | -0.127 |
| nid2a | 0.224 | pvalb1 | -0.125 |
| her11 | 0.224 | gapdh | -0.125 |
| ism1 | 0.222 | eno1a | -0.124 |
| mespaa | 0.210 | pvalb2 | -0.123 |
| draxin | 0.209 | tmem38a | -0.123 |
| zic3 | 0.208 | tnnc2 | -0.122 |
| mespab | 0.206 | ndrg2 | -0.122 |
| foxc1a | 0.203 | aldoab | -0.121 |
| arl4aa | 0.202 | mylz3 | -0.120 |
| her7 | 0.201 | neb | -0.120 |
| foxc1b | 0.199 | acta1b | -0.120 |
| BX001014.2 | 0.198 | tpi1b | -0.119 |
| si:ch73-281n10.2 | 0.193 | eif4a1b | -0.118 |
| rem1 | 0.191 | si:ch73-367p23.2 | -0.118 |
| cx43.4 | 0.190 | eno3 | -0.118 |
| zfand5a | 0.189 | ldb3b | -0.117 |
| zbtb18 | 0.188 | tnnt3a | -0.115 |
| CT030188.1 | 0.188 | nme2b.2 | -0.114 |
| apoeb | 0.183 | pmp22b | -0.113 |
| dlc | 0.182 | actn3a | -0.113 |
| alpi.1 | 0.181 | gamt | -0.112 |
| tspan7 | 0.180 | sparc | -0.112 |
| aplnra | 0.179 | srl | -0.112 |
| BX927258.1 | 0.178 | CABZ01078594.1 | -0.111 |