fibroblast growth factor 17
ZFIN 
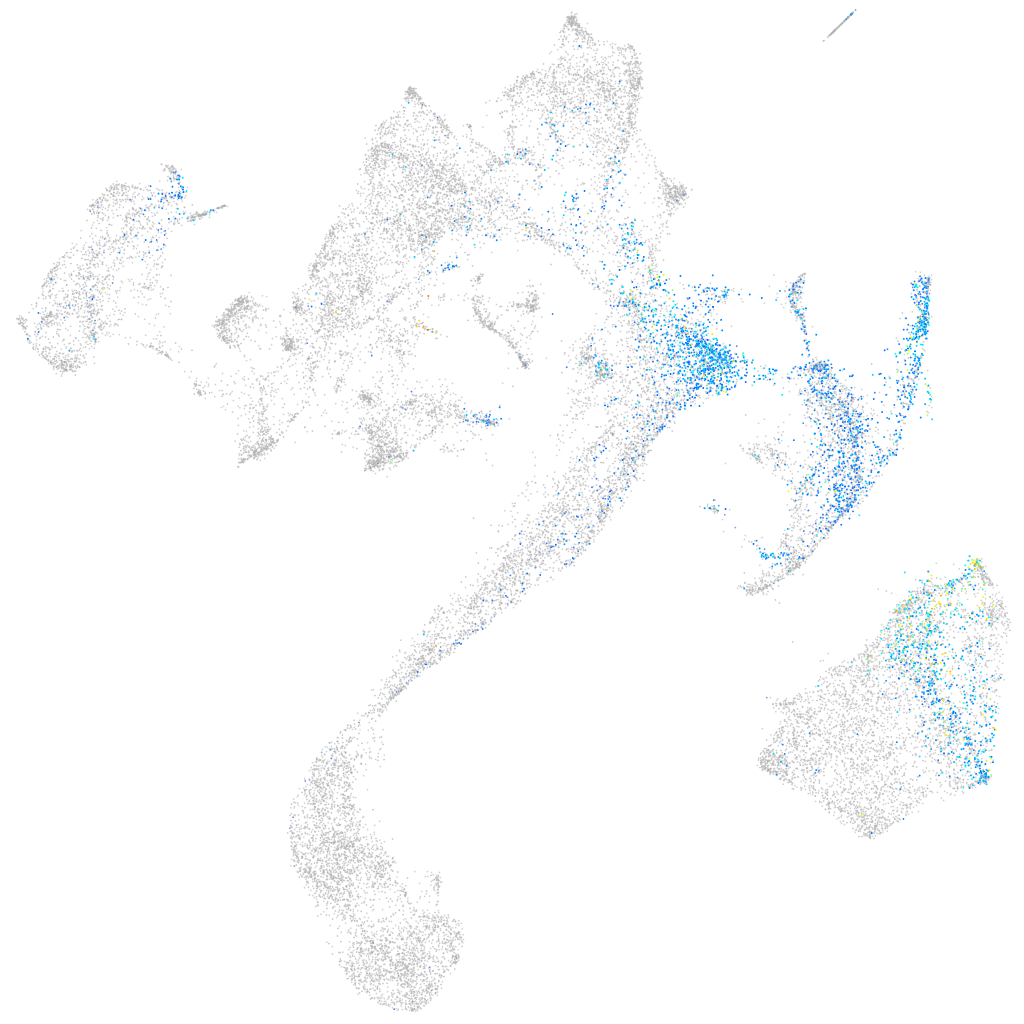
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fn1b | 0.332 | fabp3 | -0.205 |
| ripply1 | 0.327 | bhmt | -0.200 |
| kazald2 | 0.279 | ckmb | -0.182 |
| nid2a | 0.274 | atp2a1 | -0.182 |
| tjp2b | 0.273 | ckma | -0.182 |
| asph | 0.272 | mylpfa | -0.175 |
| draxin | 0.266 | pvalb1 | -0.173 |
| myf5 | 0.266 | actc1b | -0.172 |
| BX001014.2 | 0.263 | eef1da | -0.171 |
| tuba8l2 | 0.257 | pabpc4 | -0.170 |
| efnb2b | 0.256 | gamt | -0.168 |
| XLOC-042229 | 0.251 | pvalb2 | -0.168 |
| qkia | 0.242 | myl1 | -0.166 |
| BX927258.1 | 0.241 | mylz3 | -0.163 |
| aplnra | 0.240 | si:dkey-16p21.8 | -0.162 |
| meox1 | 0.239 | eno1a | -0.158 |
| apoc1 | 0.234 | ndrg2 | -0.158 |
| has2 | 0.234 | tnnt3a | -0.158 |
| si:ch73-281n10.2 | 0.232 | sparc | -0.157 |
| cx43.4 | 0.231 | si:ch73-367p23.2 | -0.157 |
| msgn1 | 0.229 | nme2b.2 | -0.154 |
| cdh2 | 0.228 | ak1 | -0.153 |
| pcdh8 | 0.222 | ldb3b | -0.151 |
| pcdh10b | 0.219 | ank1a | -0.151 |
| tbx6 | 0.216 | tnnt3b | -0.150 |
| plp2 | 0.216 | pmp22a | -0.149 |
| XLOC-032526 | 0.216 | gapdh | -0.149 |
| gnaia | 0.216 | tmem38a | -0.148 |
| ptges3b | 0.214 | col1a2 | -0.147 |
| rasgef1ba | 0.214 | tnni2a.4 | -0.147 |
| syncrip | 0.213 | rplp2 | -0.146 |
| hmga1a | 0.212 | atp1a2a | -0.145 |
| flrt3 | 0.211 | col1a1a | -0.144 |
| id1 | 0.210 | eno3 | -0.144 |
| ebna1bp2 | 0.209 | myom1a | -0.143 |