Other cell groups


all cells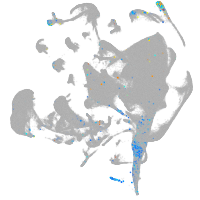 |
blastomeres |
PGCs |
endoderm |
axial mesoderm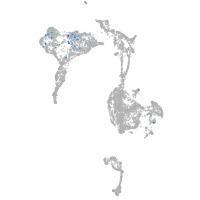 |
muscle 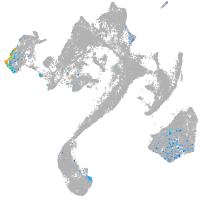 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 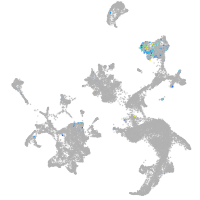 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnmb2 | 0.182 | gpm6aa | -0.058 |
| LOC110440135 | 0.174 | tuba1c | -0.058 |
| mybpc3 | 0.157 | rnasekb | -0.055 |
| zgc:195001 | 0.156 | atp6v0cb | -0.053 |
| hhatlb | 0.155 | gpm6ab | -0.052 |
| LO018250.1 | 0.151 | nova2 | -0.049 |
| myoz2b | 0.151 | stmn1b | -0.048 |
| rnf207b | 0.146 | calm1a | -0.047 |
| ryr1a | 0.145 | elavl3 | -0.046 |
| CU633479.2 | 0.145 | gng3 | -0.044 |
| chrnb1l | 0.143 | si:ch1073-429i10.3.1 | -0.044 |
| si:ch211-28p3.4 | 0.143 | cspg5a | -0.043 |
| pth1b | 0.140 | fam168a | -0.043 |
| tnnt2e | 0.139 | sncb | -0.043 |
| tnni1d | 0.136 | atpv0e2 | -0.042 |
| mpeg1.1 | 0.135 | rtn1b | -0.041 |
| havcr1 | 0.134 | vamp2 | -0.041 |
| cmklr1 | 0.132 | elavl4 | -0.040 |
| cx32.2 | 0.131 | hbbe1.3 | -0.040 |
| actn2a | 0.129 | si:dkeyp-75h12.5 | -0.040 |
| si:dkey-5n18.1 | 0.129 | marcksl1a | -0.040 |
| tnni4b.2 | 0.128 | epb41a | -0.039 |
| marco | 0.126 | stx1b | -0.039 |
| notum2 | 0.126 | ckbb | -0.038 |
| rnaset2l | 0.126 | hbae3 | -0.038 |
| itgae.1 | 0.125 | mdkb | -0.038 |
| tnni1c | 0.125 | nsg2 | -0.038 |
| LOC108180725 | 0.122 | sypb | -0.038 |
| tnnc1b | 0.122 | tubb5 | -0.038 |
| ccl34b.1 | 0.122 | atp1a3a | -0.037 |
| BX248497.1 | 0.120 | cadm3 | -0.037 |
| mustn1b | 0.120 | ccni | -0.037 |
| myh7bb | 0.120 | olfm1b | -0.037 |
| tnnt2d | 0.120 | snap25a | -0.037 |
| hspb11 | 0.118 | fez1 | -0.036 |




