"actinin, alpha 2a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 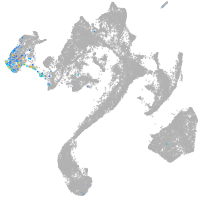 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye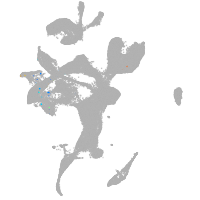 |
taste / olfactory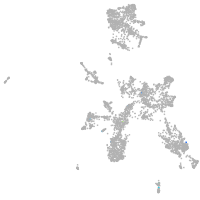 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110440135 | 0.372 | gpm6aa | -0.032 |
| kcnmb2 | 0.307 | gpm6ab | -0.029 |
| hhatlb | 0.294 | marcksl1a | -0.027 |
| mybpc3 | 0.292 | mdkb | -0.025 |
| LO018250.1 | 0.284 | nova2 | -0.025 |
| chrnb1l | 0.283 | tubb5 | -0.025 |
| rnf207b | 0.281 | cspg5a | -0.024 |
| ryr1a | 0.279 | rnasekb | -0.024 |
| myoz2b | 0.278 | rtn1b | -0.024 |
| notum2 | 0.269 | atp6v0cb | -0.023 |
| tnnt2e | 0.266 | ckbb | -0.023 |
| tnni1d | 0.264 | tuba1c | -0.023 |
| CU633479.2 | 0.263 | ywhaqb | -0.023 |
| tnni1c | 0.252 | calm3b | -0.022 |
| zgc:195001 | 0.252 | aplp1 | -0.021 |
| smyhc2 | 0.250 | atp6ap2 | -0.021 |
| si:ch211-28p3.4 | 0.246 | calr | -0.021 |
| tnnc1b | 0.242 | chd4a | -0.021 |
| tnni4b.2 | 0.242 | stmn1b | -0.021 |
| BX248497.1 | 0.239 | atp6v0ca | -0.020 |
| myh7bb | 0.238 | cd99l2 | -0.020 |
| actc1c | 0.229 | elavl3 | -0.020 |
| smyhc1 | 0.224 | fam168a | -0.020 |
| mustn1b | 0.223 | gapdhs | -0.020 |
| tnnt2d | 0.223 | nsg2 | -0.020 |
| mylk3 | 0.221 | sv2a | -0.020 |
| si:ch211-157b11.14 | 0.215 | tmeff1b | -0.020 |
| smyhc3 | 0.208 | tmem59l | -0.020 |
| mylk4a | 0.207 | zc4h2 | -0.020 |
| myom1b | 0.206 | appb | -0.019 |
| pvalb4 | 0.205 | elavl4 | -0.019 |
| myl13 | 0.204 | nrxn1a | -0.019 |
| myl10 | 0.203 | olfm1b | -0.019 |
| si:dkeyp-68b7.10 | 0.199 | si:ch211-288g17.3 | -0.019 |
| sbk3 | 0.195 | stx1b | -0.019 |




