ectonucleotide pyrophosphatase/phosphodiesterase 2
ZFIN 
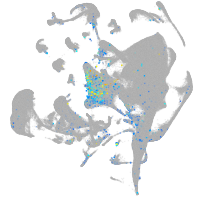
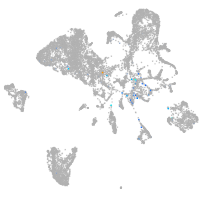
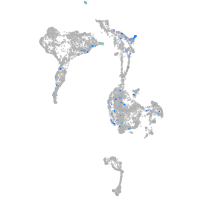
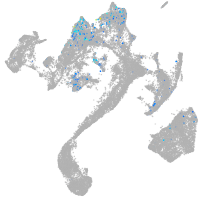
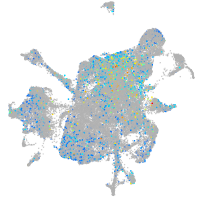
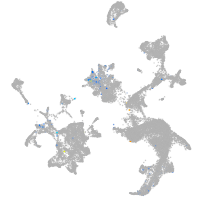
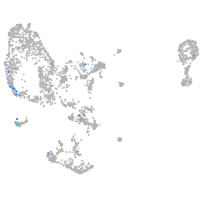
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| loxl5b | 0.187 | hmgb1b | -0.039 |
| clu | 0.183 | ptmab | -0.039 |
| hpgd | 0.182 | hnrnpaba | -0.036 |
| slit1b | 0.177 | khdrbs1a | -0.035 |
| myl9a | 0.154 | si:ch211-288g17.3 | -0.035 |
| ptgs2a | 0.147 | marcksb | -0.034 |
| vasna | 0.143 | tubb2b | -0.032 |
| shhb | 0.143 | nova2 | -0.032 |
| spon1b | 0.139 | elavl3 | -0.032 |
| npr3 | 0.137 | apex1 | -0.032 |
| LOC108181595 | 0.135 | hmgb2b | -0.032 |
| arxa | 0.131 | h3f3a | -0.031 |
| CU467042.1 | 0.128 | hsp90ab1 | -0.031 |
| tmem88b | 0.127 | tmeff1b | -0.030 |
| LOC108179692 | 0.126 | hnrnpa0b | -0.030 |
| LOC110439500 | 0.124 | seta | -0.029 |
| slit2 | 0.121 | sox11b | -0.028 |
| si:dkey-208c12.2 | 0.121 | si:dkey-276j7.1 | -0.028 |
| emp2 | 0.121 | hmgb3a | -0.028 |
| si:ch211-241f5.3 | 0.120 | myt1b | -0.028 |
| trabd2b | 0.120 | si:ch73-1a9.3 | -0.028 |
| abcc6b.1 | 0.119 | cfl1 | -0.028 |
| foxa1 | 0.116 | hmgn2 | -0.028 |
| shha | 0.116 | nono | -0.028 |
| sparcl1 | 0.115 | syncrip | -0.028 |
| ctgfa | 0.114 | hmga1a | -0.027 |
| zgc:162730 | 0.113 | her4.2 | -0.027 |
| urp2 | 0.112 | mdkb | -0.027 |
| col5a1 | 0.109 | rtn1a | -0.027 |
| LOC110438075 | 0.109 | cirbpb | -0.027 |
| palm3 | 0.108 | sox3 | -0.027 |
| XLOC-005056 | 0.107 | setb | -0.027 |
| bmp1a | 0.106 | nkain1 | -0.026 |
| olfml2ba | 0.106 | sinhcafl | -0.026 |
| ntn1b | 0.105 | hnrnph1l | -0.026 |