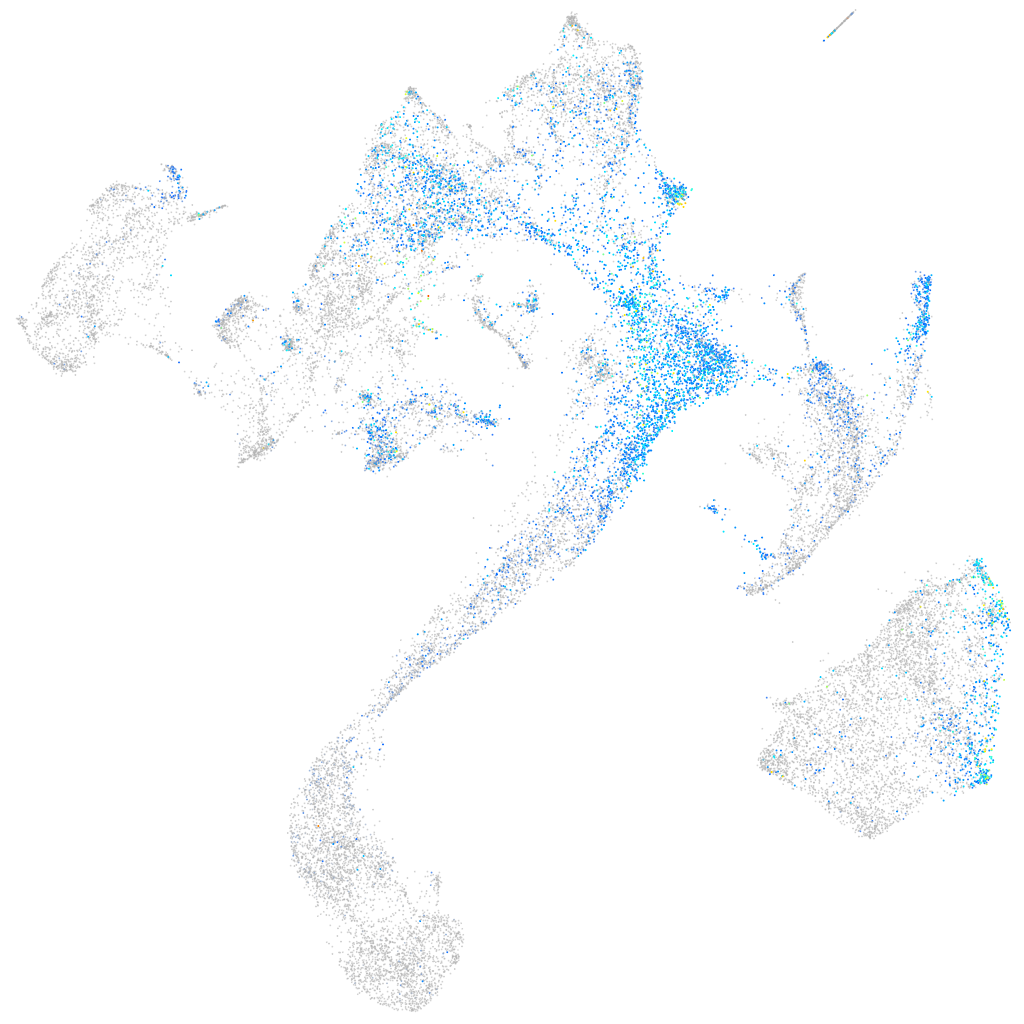
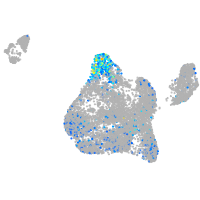
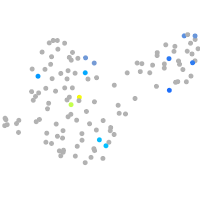
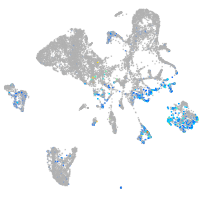
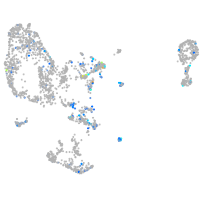
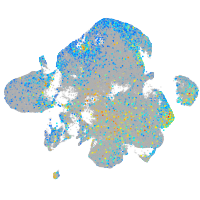
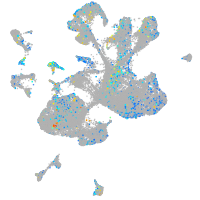
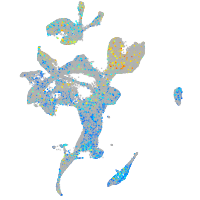
ephrin-B2a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| uncx4.1 | 0.366 | ckma | -0.246 |
| cpn1 | 0.336 | ckmb | -0.242 |
| tcf15 | 0.323 | atp2a1 | -0.237 |
| kirrel3l | 0.323 | gapdh | -0.226 |
| angptl7 | 0.322 | actc1b | -0.223 |
| meox1 | 0.322 | si:ch73-367p23.2 | -0.216 |
| mdka | 0.319 | tnnc2 | -0.215 |
| XLOC-042229 | 0.319 | aldoab | -0.215 |
| emp2 | 0.306 | ak1 | -0.214 |
| nr2f5 | 0.300 | ldb3b | -0.213 |
| fn1b | 0.294 | mylpfa | -0.212 |
| aldh1a2 | 0.291 | tnnt3a | -0.211 |
| rdh10a | 0.290 | neb | -0.210 |
| rprmb | 0.285 | eno3 | -0.210 |
| meis1a | 0.276 | ank1a | -0.208 |
| bmpr1ba | 0.274 | mylz3 | -0.207 |
| id1 | 0.272 | tmem38a | -0.206 |
| fat2 | 0.269 | myom1a | -0.205 |
| ripply1 | 0.268 | actn3a | -0.205 |
| flrt3 | 0.265 | pgam2 | -0.204 |
| optc | 0.261 | pvalb1 | -0.203 |
| kazald2 | 0.260 | pvalb2 | -0.201 |
| mfap2 | 0.259 | si:ch211-266g18.10 | -0.200 |
| dmrt2a | 0.259 | smyd1a | -0.200 |
| alcama | 0.255 | acta1b | -0.198 |
| btg2 | 0.255 | prx | -0.198 |
| pax3a | 0.254 | nme2b.2 | -0.196 |
| rasgef1ba | 0.252 | tnnt3b | -0.195 |
| her6 | 0.250 | hhatla | -0.194 |
| efemp2a | 0.247 | XLOC-025819 | -0.193 |
| thbs4b | 0.247 | cavin4a | -0.193 |
| ssr3 | 0.244 | casq2 | -0.191 |
| ran | 0.242 | tmod4 | -0.191 |
| hsp90ab1 | 0.242 | casq1b | -0.191 |
| naca | 0.242 | tnni2a.4 | -0.189 |