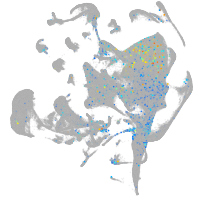
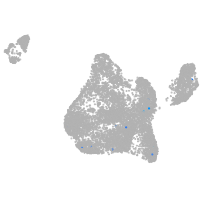
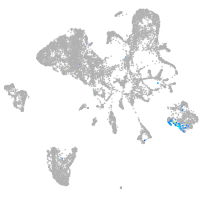
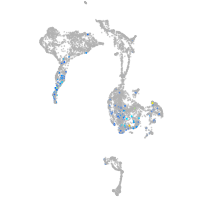
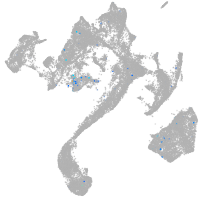
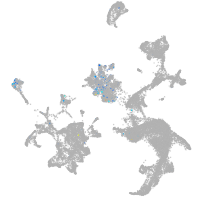
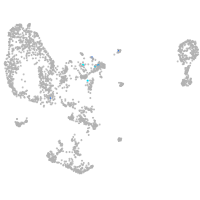
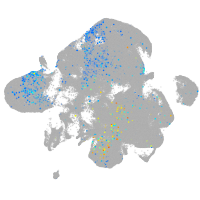
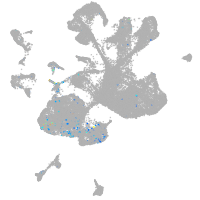
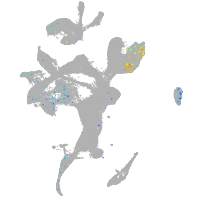
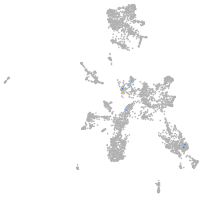
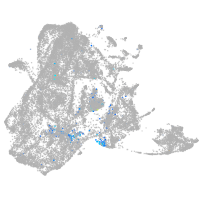
diencephalon/mesencephalon homeobox 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp26c1 | 0.527 | rps10 | -0.196 |
| otx1 | 0.461 | rpl37 | -0.195 |
| gsc | 0.384 | zgc:114188 | -0.183 |
| si:dkey-43p13.5 | 0.337 | rps17 | -0.165 |
| otx2b | 0.319 | nme2b.1 | -0.138 |
| si:dkey-88l16.5 | 0.300 | zgc:56493 | -0.119 |
| nog1 | 0.299 | ahcy | -0.105 |
| iqca1 | 0.280 | eef1da | -0.104 |
| shisa2a | 0.268 | prdx2 | -0.103 |
| her5 | 0.255 | zgc:92744 | -0.100 |
| otx2a | 0.254 | eef1b2 | -0.100 |
| fzd8b | 0.251 | atp5if1b | -0.099 |
| si:ch211-152c2.3 | 0.236 | COX7A2 (1 of many) | -0.098 |
| CU571091.1 | 0.232 | eno3 | -0.097 |
| gpm6ab | 0.228 | cox6a1 | -0.096 |
| BX649529.1 | 0.227 | zgc:158463 | -0.095 |
| akap12b | 0.227 | atp5pf | -0.095 |
| pltp | 0.227 | atp5mc3b | -0.093 |
| dbx1a | 0.226 | atp5f1b | -0.092 |
| cxcl12b | 0.223 | mt-nd1 | -0.092 |
| asph | 0.221 | suclg1 | -0.091 |
| zgc:162707 | 0.220 | rps20 | -0.091 |
| nes | 0.220 | sod1 | -0.091 |
| lft2 | 0.218 | atp5l | -0.090 |
| pkdcca | 0.217 | gapdh | -0.089 |
| marcksl1b | 0.214 | rpl9 | -0.089 |
| fscn1a | 0.214 | dap | -0.087 |
| sfrp1a | 0.211 | gstp1 | -0.085 |
| grhl2b | 0.207 | atp5fa1 | -0.085 |
| BX120006.3 | 0.205 | gamt | -0.085 |
| cdh6 | 0.204 | rplp0 | -0.085 |
| arhgap36 | 0.202 | rps14 | -0.084 |
| dhrs3b | 0.192 | cox5aa | -0.084 |
| flrt3 | 0.189 | atp5mc1 | -0.084 |
| lima1a | 0.189 | rpl19 | -0.082 |