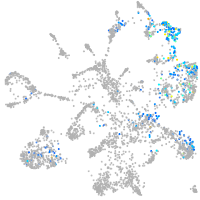
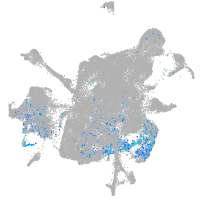
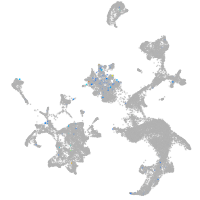
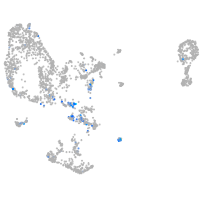
crumbs cell polarity complex component 2a
ZFIN 
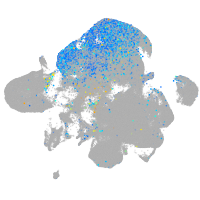
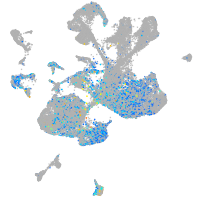
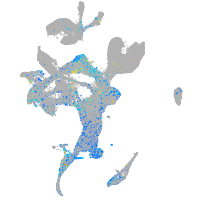
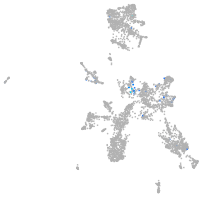
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msi1 | 0.253 | aldob | -0.065 |
| otx5 | 0.222 | gapdh | -0.065 |
| XLOC-016325 | 0.212 | gamt | -0.058 |
| prdm1b | 0.201 | eno3 | -0.056 |
| zgc:110045 | 0.198 | glud1b | -0.054 |
| si:ch211-202e12.3 | 0.193 | fbp1b | -0.053 |
| pde6gb | 0.191 | ckba | -0.053 |
| XLOC-007573 | 0.177 | cebpd | -0.053 |
| proca1 | 0.176 | eef1da | -0.053 |
| cldn5a | 0.168 | tpi1b | -0.053 |
| LOC101884182 | 0.167 | BX908782.3 | -0.052 |
| crx | 0.166 | sod1 | -0.052 |
| AL954142.2 | 0.159 | pklr | -0.052 |
| slc2a15a | 0.157 | nupr1b | -0.052 |
| si:ch211-284e13.14 | 0.155 | atp5if1b | -0.050 |
| XLOC-017089 | 0.155 | dap | -0.050 |
| lrtm1 | 0.153 | mdh1aa | -0.049 |
| zic6 | 0.150 | cx32.3 | -0.049 |
| pkd1l1 | 0.147 | agxtb | -0.049 |
| CU651662.1 | 0.147 | gnmt | -0.048 |
| XLOC-003690 | 0.146 | abat | -0.048 |
| BX470117.1 | 0.142 | sod2 | -0.048 |
| fabp7a | 0.140 | pnp4b | -0.048 |
| gsg1l2b | 0.139 | scp2a | -0.047 |
| LOC108181556 | 0.137 | apoa4b.1 | -0.047 |
| XLOC-027134 | 0.136 | suclg1 | -0.046 |
| ecm2 | 0.136 | cx28.9 | -0.046 |
| vtg5 | 0.135 | pgk1 | -0.045 |
| mdka | 0.134 | fabp10a | -0.045 |
| nfe2 | 0.133 | cat | -0.045 |
| neurod4 | 0.130 | srd5a2a | -0.045 |
| plp1a | 0.130 | apoc2 | -0.045 |
| cspg5b | 0.130 | pgm1 | -0.044 |
| LOC108191536 | 0.129 | suclg2 | -0.044 |
| LOC555502 | 0.128 | gstp1 | -0.044 |