"phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b"
ZFIN 
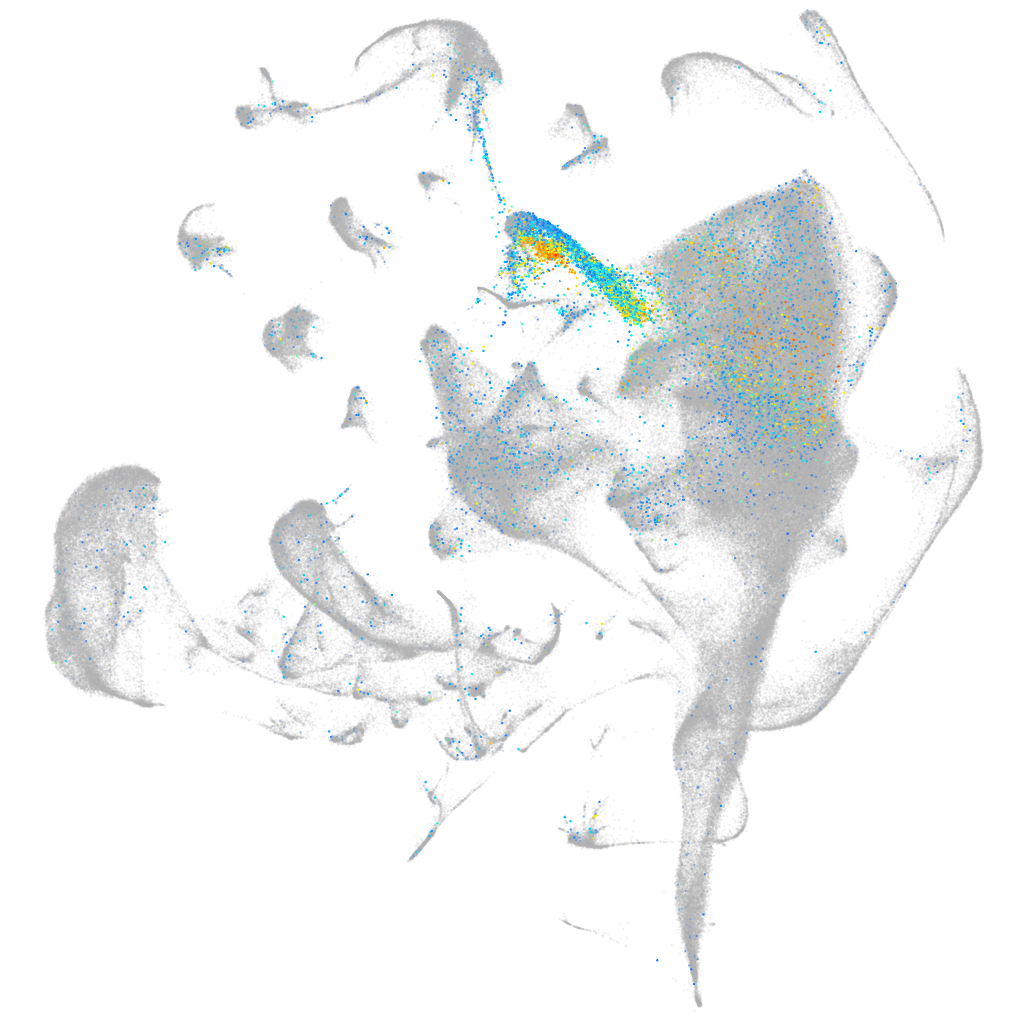
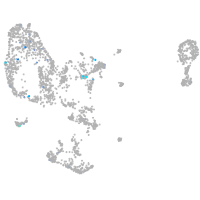
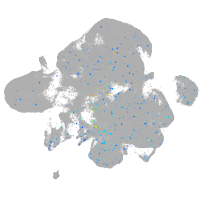
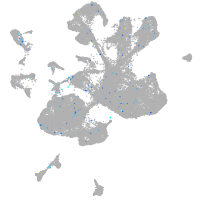
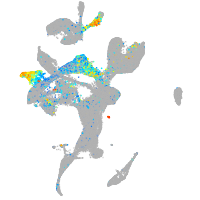
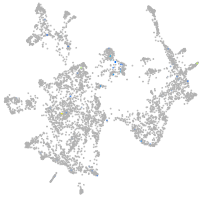
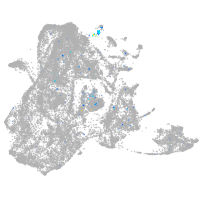
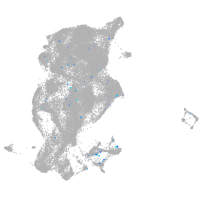
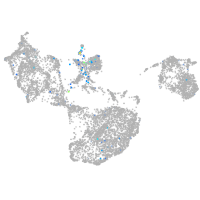
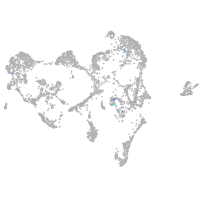
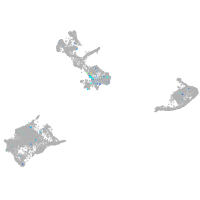
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nr2e3 | 0.448 | id1 | -0.086 |
| si:dkey-21o19.2 | 0.428 | fosab | -0.081 |
| cnga1a | 0.375 | bzw1b | -0.075 |
| cnga1b | 0.371 | ctsla | -0.071 |
| otx5 | 0.371 | vamp3 | -0.071 |
| rom1b | 0.369 | eif4ebp3l | -0.069 |
| saga | 0.362 | btg2 | -0.067 |
| sagb | 0.349 | aldob | -0.066 |
| gngt2a | 0.344 | rps27l | -0.063 |
| gnat1 | 0.340 | zfp36l1a | -0.063 |
| si:ch211-113d22.2 | 0.340 | arhgdia | -0.062 |
| zgc:109965 | 0.335 | aldh9a1a.1 | -0.059 |
| elovl4b | 0.334 | her6 | -0.059 |
| crx | 0.333 | junba | -0.059 |
| rhol | 0.331 | qkia | -0.059 |
| XLOC-014477 | 0.330 | slc38a5b | -0.059 |
| rom1a | 0.325 | sparc | -0.059 |
| si:ch73-28h20.1 | 0.321 | ctnnb1 | -0.058 |
| samd7 | 0.320 | ier2b | -0.058 |
| guca1b | 0.315 | tpm4a | -0.057 |
| prdm1b | 0.310 | cd63 | -0.056 |
| BX004774.2 | 0.302 | lasp1 | -0.056 |
| gngt1 | 0.296 | tpm3 | -0.056 |
| grk1a | 0.296 | wls | -0.056 |
| zgc:112294 | 0.296 | flot1b | -0.055 |
| tmx3a | 0.294 | foxp4 | -0.055 |
| pde6ga | 0.284 | GCA | -0.055 |
| kcnv2a | 0.281 | her9 | -0.055 |
| CR361564.1 | 0.278 | krt8 | -0.055 |
| arhgap20 | 0.277 | tuba8l2 | -0.055 |
| pde6b | 0.268 | gsta.1 | -0.054 |
| tmem244 | 0.268 | ier2a | -0.054 |
| faimb | 0.262 | XLOC-044191 | -0.054 |
| neurod1 | 0.260 | zgc:162730 | -0.053 |
| nrl | 0.258 | ahnak | -0.052 |