crumbs cell polarity complex component 2a
ZFIN 
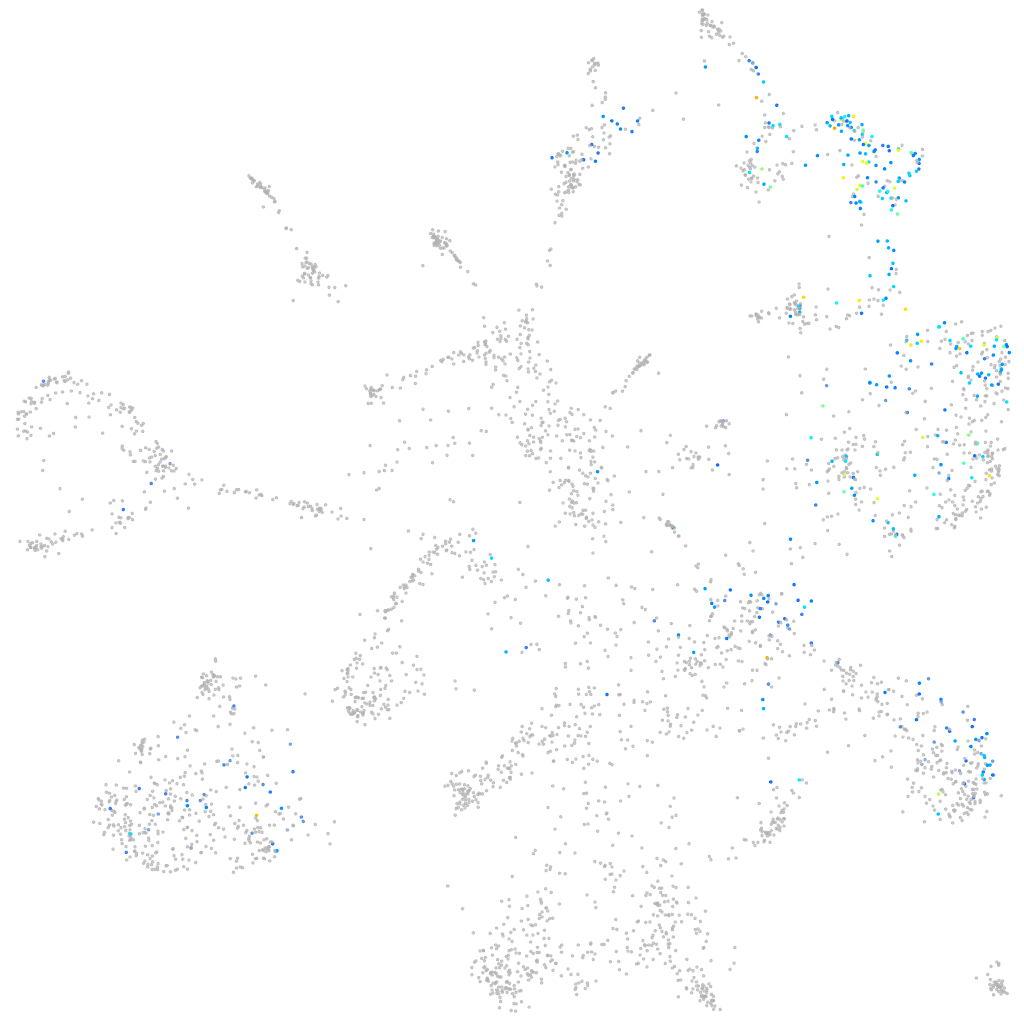
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sfrp5 | 0.411 | BX088707.3 | -0.227 |
| tmem88b | 0.357 | tpm1 | -0.222 |
| jam2b | 0.341 | myl9a | -0.209 |
| podxl | 0.339 | acta2 | -0.194 |
| CABZ01075068.1 | 0.321 | tagln | -0.190 |
| COLEC10 | 0.321 | myh11a | -0.172 |
| synpo2lb | 0.317 | mylkb | -0.165 |
| alpi.1 | 0.307 | ptmaa | -0.160 |
| cd151 | 0.305 | lmod1b | -0.155 |
| angptl6 | 0.302 | gapdhs | -0.142 |
| cxadr | 0.298 | myl6 | -0.141 |
| mmel1 | 0.291 | tpm2 | -0.141 |
| gata5 | 0.290 | col4a1 | -0.136 |
| XLOC-042222 | 0.290 | si:ch211-62a1.3 | -0.136 |
| ftr82 | 0.289 | inka1a | -0.135 |
| nr0b2a | 0.287 | fhl2a | -0.134 |
| fabp11a | 0.285 | csrp1b | -0.132 |
| slc29a1b | 0.279 | XLOC-025423 | -0.128 |
| tnni1b | 0.277 | ckbb | -0.126 |
| alx4a | 0.275 | zgc:153704 | -0.124 |
| krt94 | 0.271 | ak1 | -0.124 |
| lmod2a | 0.269 | desmb | -0.122 |
| krt18a.1 | 0.267 | cnn1b | -0.121 |
| gata6 | 0.261 | igfbp7 | -0.120 |
| krt8 | 0.261 | cald1b | -0.120 |
| si:ch211-214p16.2 | 0.258 | mylka | -0.118 |
| tmem88a | 0.255 | mcamb | -0.117 |
| thy1 | 0.255 | BX323087.1 | -0.112 |
| tbx5a | 0.254 | myocd | -0.108 |
| cx43.4 | 0.254 | col4a2 | -0.108 |
| akap12b | 0.252 | pdgfrb | -0.107 |
| tmem108 | 0.246 | foxf1 | -0.107 |
| mdka | 0.246 | calm2a | -0.106 |
| tgm2b | 0.245 | ppiab | -0.105 |
| hspb1 | 0.244 | ntn5 | -0.105 |