CKLF-like MARVEL transmembrane domain containing 4
ZFIN 
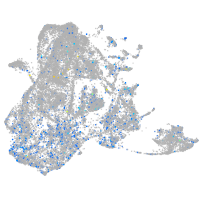
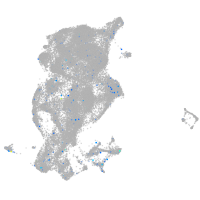
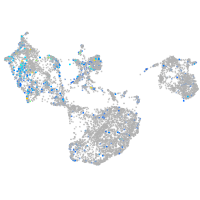
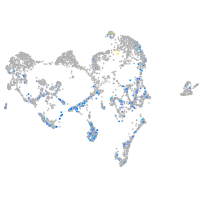
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gcm2 | 0.265 | krt91 | -0.176 |
| etf1a | 0.251 | atp1a1a.2 | -0.139 |
| atp6v0a1a | 0.246 | si:dkey-193p11.2 | -0.139 |
| slc6a6b | 0.239 | si:dkey-189h5.6 | -0.128 |
| foxi3a | 0.238 | BX000438.2 | -0.126 |
| atp6v1ba | 0.236 | icn2 | -0.125 |
| ca15a | 0.235 | krt5 | -0.118 |
| atp6v1c1b | 0.233 | slc12a10.2 | -0.118 |
| CABZ01030107.1 | 0.231 | krt17 | -0.117 |
| atp6ap1b | 0.230 | clcn2c | -0.117 |
| atp6v1aa | 0.228 | krtt1c19e | -0.115 |
| si:dkey-192d15.2 | 0.226 | si:dkey-184p9.7 | -0.114 |
| ceacam1 | 0.225 | atp1a1a.3 | -0.113 |
| atp6v0ca | 0.225 | slc4a4b | -0.108 |
| atp6v1e1b | 0.224 | prlra | -0.105 |
| atp6v1g1 | 0.223 | lgals3b | -0.105 |
| atp6ap2 | 0.223 | pcdh11 | -0.104 |
| CDK18 | 0.220 | malb | -0.104 |
| atp6v0d1 | 0.215 | mgst1.2 | -0.102 |
| cox5b2 | 0.212 | egr2a | -0.101 |
| atp6v1d | 0.209 | gsto1 | -0.100 |
| slc41a1 | 0.208 | aldob | -0.100 |
| rnaseka | 0.207 | ndrg1a | -0.100 |
| zgc:193726 | 0.205 | nudt4a | -0.099 |
| ca2 | 0.201 | krt4 | -0.098 |
| slc31a2 | 0.200 | mef2aa | -0.096 |
| atp6v0b | 0.200 | pycard | -0.095 |
| rpia | 0.199 | zgc:163083 | -0.093 |
| lsp1 | 0.198 | rnf183 | -0.093 |
| ppp1r3ab | 0.198 | si:ch73-359m17.9 | -0.093 |
| slc6a6a | 0.198 | anxa2a | -0.092 |
| slc4a1b | 0.198 | pnp5a | -0.090 |
| clcn2b | 0.193 | agr1 | -0.090 |
| tfe3b | 0.192 | foxq1a | -0.090 |
| arhgap32a | 0.189 | rplp2 | -0.089 |