CDC42 effector protein (Rho GTPase binding) 1a
ZFIN 
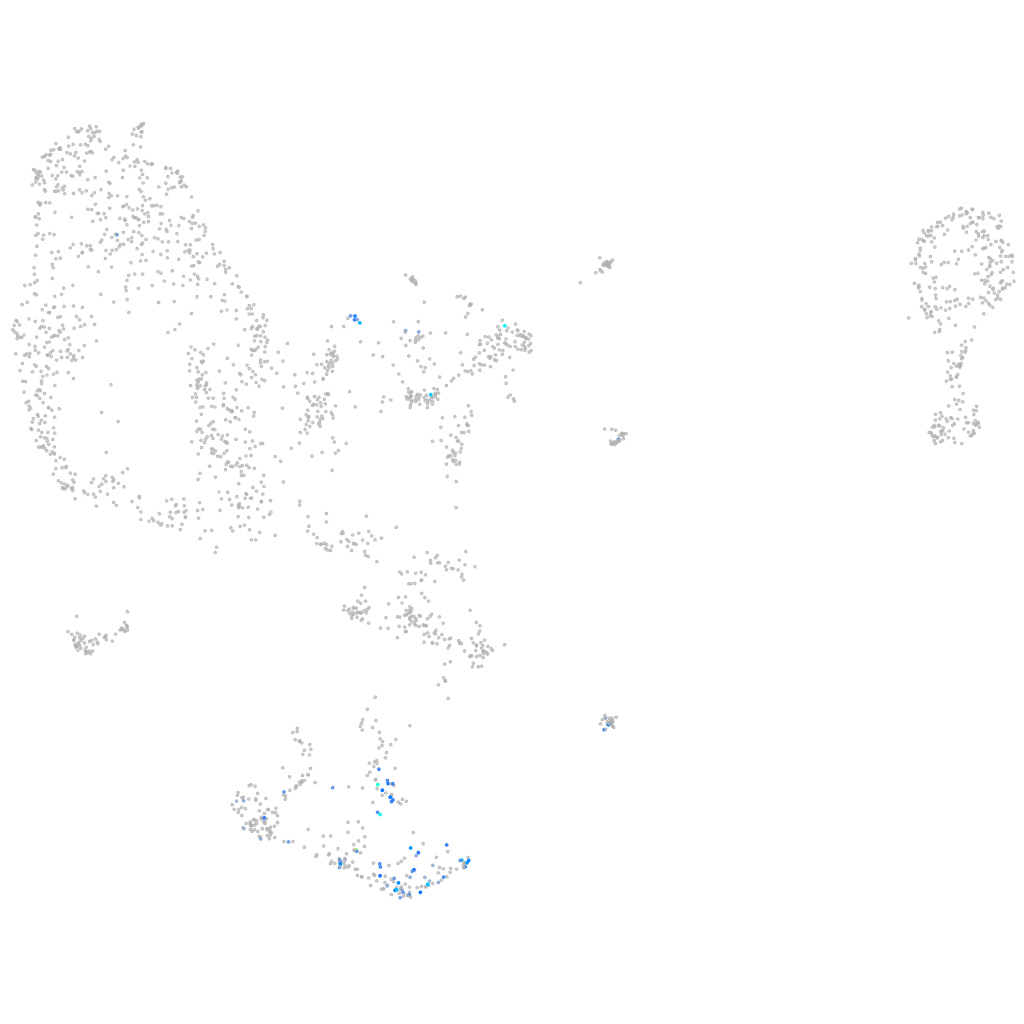
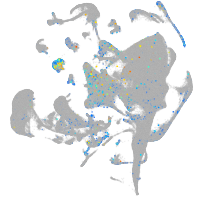
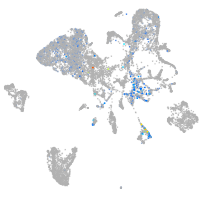
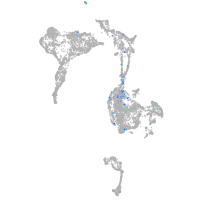
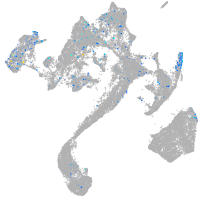
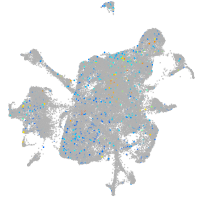
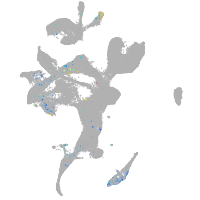
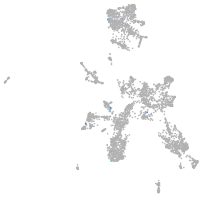
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-36i7.3 | 0.372 | acy3.2 | -0.153 |
| ppp1r1b | 0.356 | si:rp71-17i16.6 | -0.125 |
| soul2 | 0.344 | si:dkey-194e6.1 | -0.118 |
| trim35-12 | 0.320 | aqp8b | -0.116 |
| atp1a1a.3 | 0.319 | pdzk1 | -0.112 |
| tmem176l.4 | 0.318 | grhprb | -0.111 |
| gpa33a | 0.315 | slc9a3r1a | -0.108 |
| rnf183 | 0.312 | eif5a2 | -0.107 |
| atp1a1a.5 | 0.310 | slc26a1 | -0.106 |
| slc12a10.3 | 0.304 | slc4a4a | -0.105 |
| me3 | 0.300 | ftcd | -0.104 |
| znf1070 | 0.298 | zgc:101040 | -0.104 |
| gdpd3a | 0.298 | pdzk1ip1 | -0.104 |
| angpt2a | 0.296 | hao1 | -0.104 |
| ehd4 | 0.289 | lrpap1 | -0.103 |
| quo | 0.288 | slc26a6 | -0.103 |
| AL954322.2 | 0.288 | dab2 | -0.102 |
| tbx2b | 0.286 | gamt | -0.100 |
| oclnb | 0.286 | upb1 | -0.100 |
| klf5l | 0.285 | lrp2a | -0.100 |
| ctgfa | 0.283 | serbp1a | -0.099 |
| clcnk | 0.282 | gnpda1 | -0.099 |
| fabp2 | 0.279 | fbp1b | -0.099 |
| si:ch73-359m17.9 | 0.278 | pnp6 | -0.098 |
| thbs1b | 0.277 | ahcyl1 | -0.098 |
| mctp2b | 0.273 | nit2 | -0.098 |
| etf1a | 0.271 | syncrip | -0.097 |
| slc4a5a | 0.271 | agxta | -0.096 |
| BX000438.2 | 0.270 | gstt1a | -0.095 |
| hoxc11b | 0.269 | cubn | -0.095 |
| plecb | 0.268 | gstr | -0.095 |
| mal2 | 0.267 | si:dkey-28n18.9 | -0.095 |
| p2ry2.1 | 0.265 | ptges3b | -0.094 |
| phlda2 | 0.264 | snrpf | -0.094 |
| phkg1b | 0.260 | npr1b | -0.094 |