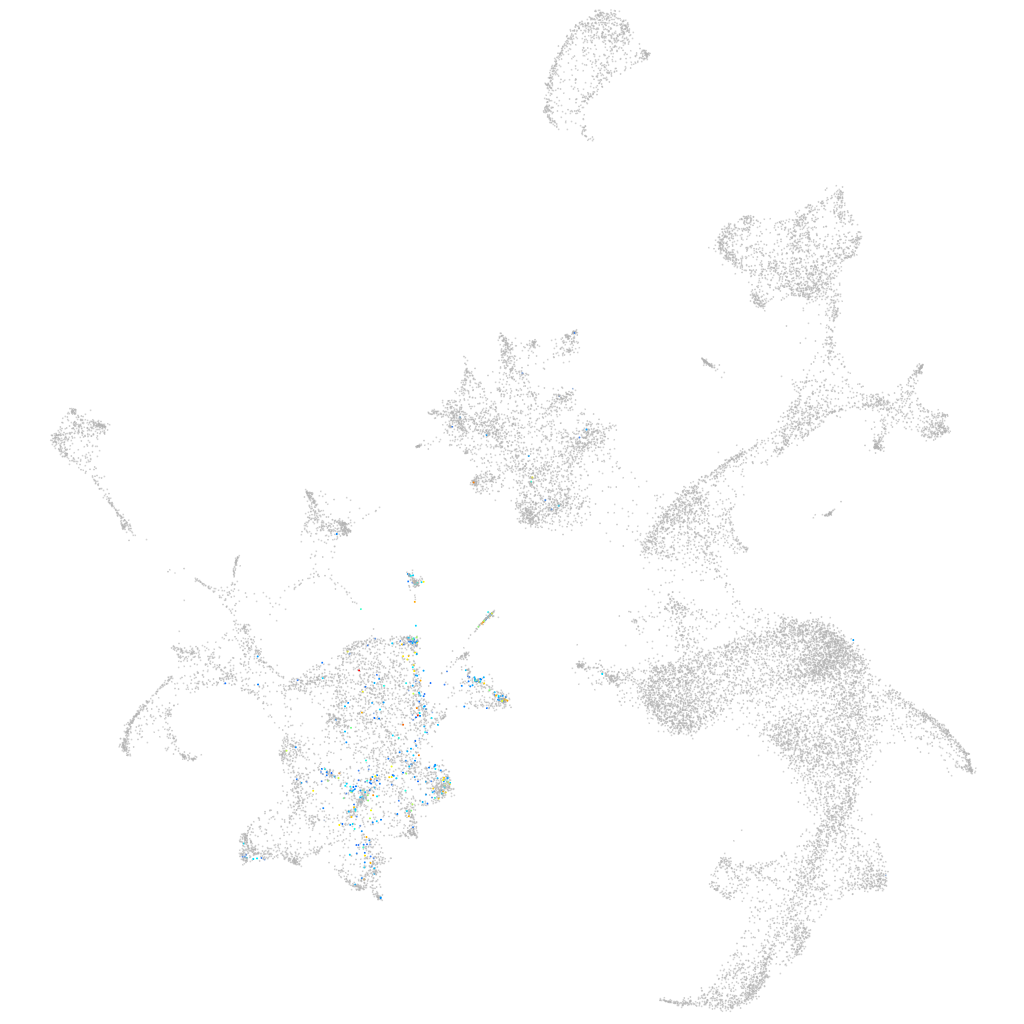
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cpn1 | 0.235 | hbae1.1 | -0.113 |
| plvapb | 0.202 | prdx2 | -0.109 |
| igfbp7 | 0.197 | hbbe1.3 | -0.106 |
| ramp2 | 0.190 | hbae1.3 | -0.105 |
| myct1a | 0.190 | hbbe2 | -0.105 |
| angpt2a | 0.187 | hbae3 | -0.105 |
| adma | 0.185 | hbbe1.1 | -0.101 |
| ednraa | 0.185 | cahz | -0.099 |
| sox18 | 0.185 | fth1a | -0.096 |
| pcdh12 | 0.184 | hbbe1.2 | -0.093 |
| cldn5b | 0.180 | blvrb | -0.093 |
| fam174b | 0.180 | hemgn | -0.092 |
| plk2b | 0.179 | alas2 | -0.087 |
| tgfbr2b | 0.179 | si:ch211-250g4.3 | -0.083 |
| grb10a | 0.179 | slc4a1a | -0.082 |
| cd81b | 0.176 | nt5c2l1 | -0.080 |
| ecscr | 0.176 | gpx1a | -0.078 |
| krt18a.1 | 0.175 | tspo | -0.078 |
| krt8 | 0.174 | epb41b | -0.077 |
| si:dkeyp-97a10.2 | 0.174 | zgc:163057 | -0.077 |
| plk2a | 0.172 | creg1 | -0.074 |
| exoc3l2a | 0.171 | si:ch211-207c6.2 | -0.073 |
| tspan4b | 0.171 | nmt1b | -0.070 |
| vat1 | 0.169 | plac8l1 | -0.068 |
| hlx1 | 0.169 | tmod4 | -0.065 |
| lyve1a | 0.168 | znfl2a | -0.062 |
| egfl7 | 0.168 | hbae5 | -0.061 |
| rgcc | 0.166 | mibp2 | -0.060 |
| fxyd6l | 0.166 | tdh | -0.059 |
| pecam1 | 0.165 | hdr | -0.059 |
| notchl | 0.163 | tfr1a | -0.059 |
| tmem108 | 0.163 | hbbe3 | -0.059 |
| plekhg5a | 0.161 | sptb | -0.059 |
| dusp5 | 0.161 | zgc:56095 | -0.059 |
| si:ch211-145b13.6 | 0.161 | rfesd | -0.058 |