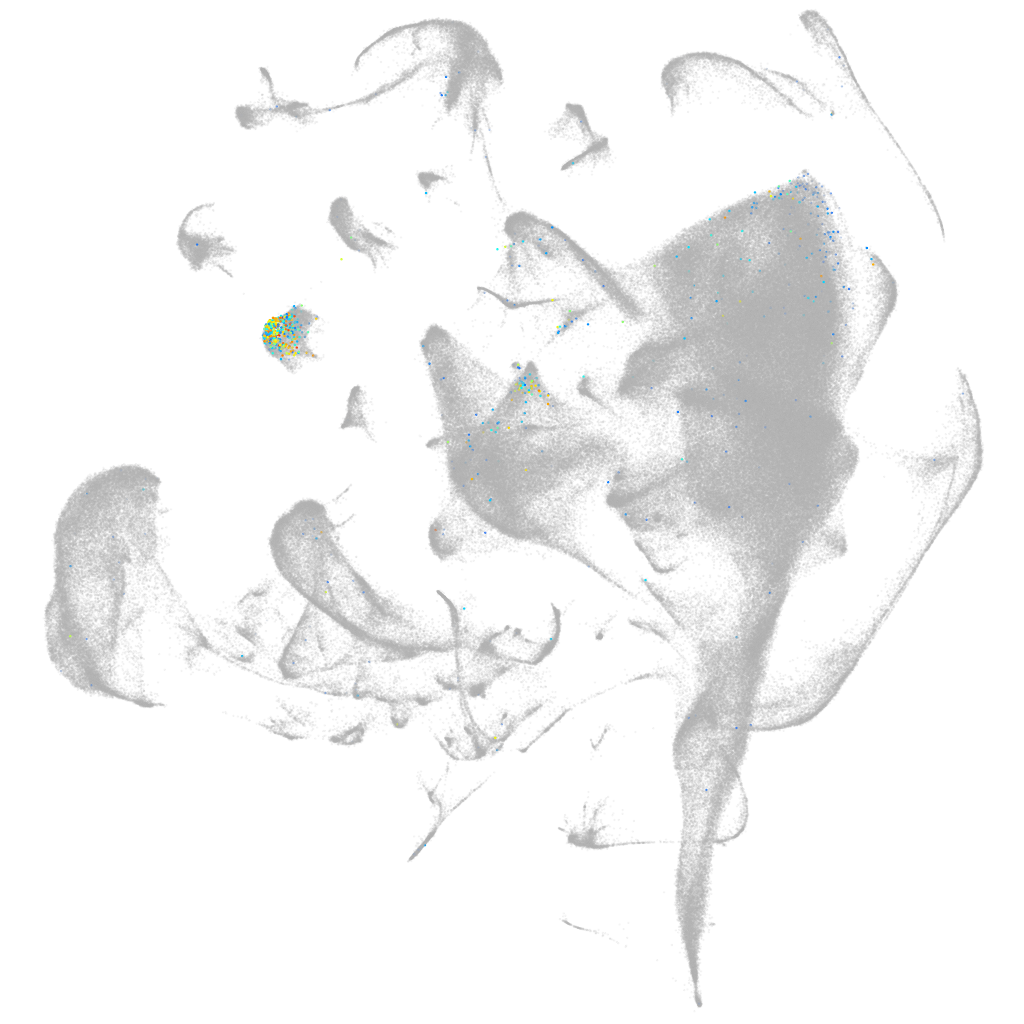
lymphatic vessel endothelial hyaluronic receptor 1a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 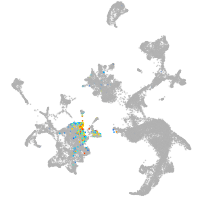 |
pronephros 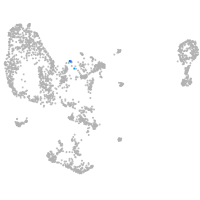 |
neural |
spinal cord / glia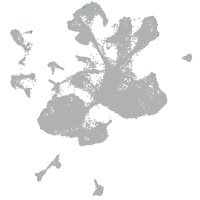 |
eye |
taste / olfactory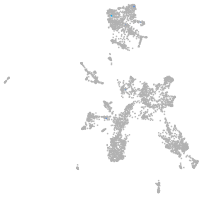 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lyve1b | 0.324 | fabp3 | -0.026 |
| gpr182 | 0.277 | cx43.4 | -0.025 |
| mrc1a | 0.271 | marcksb | -0.024 |
| stab1 | 0.258 | hmgb1b | -0.023 |
| plvapb | 0.252 | si:ch211-137a8.4 | -0.023 |
| sele | 0.246 | si:ch211-288g17.3 | -0.023 |
| si:ch211-145b13.6 | 0.246 | ddx39ab | -0.022 |
| si:ch211-33e4.2 | 0.231 | snrpb | -0.022 |
| tie1 | 0.229 | nucks1a | -0.021 |
| stab2 | 0.205 | snrpe | -0.021 |
| exoc3l2a | 0.204 | hnrnpa1b | -0.020 |
| pecam1 | 0.198 | seta | -0.020 |
| clec14a | 0.195 | snrpd1 | -0.020 |
| XLOC-014079 | 0.195 | ncl | -0.019 |
| myct1a | 0.193 | ptges3b | -0.019 |
| etv2 | 0.191 | setb | -0.019 |
| kdr | 0.191 | snrpf | -0.019 |
| dab2 | 0.189 | apex1 | -0.018 |
| si:dkey-28n18.9 | 0.187 | ddx39aa | -0.018 |
| si:ch73-334d15.2 | 0.183 | hnrnph1l | -0.018 |
| si:dkeyp-97a10.2 | 0.179 | magoh | -0.018 |
| srgn | 0.176 | rbbp4 | -0.018 |
| XLOC-039757 | 0.176 | snu13b | -0.018 |
| cdh5 | 0.175 | sox3 | -0.018 |
| ecscr | 0.175 | stmn1a | -0.018 |
| XLOC-043043 | 0.170 | her15.1 | -0.018 |
| ehd4 | 0.165 | alyref | -0.017 |
| calcrlb | 0.163 | dkc1 | -0.017 |
| pcdh12 | 0.161 | gpm6aa | -0.017 |
| si:ch211-14k19.8 | 0.161 | hnrnpub | -0.017 |
| mrc1b | 0.156 | ilf2 | -0.017 |
| fgd5a | 0.155 | lig1 | -0.017 |
| kdrl | 0.155 | snrpd2 | -0.017 |
| ramp2 | 0.155 | srsf3b | -0.017 |
| si:ch211-66e2.3 | 0.155 | ssrp1a | -0.017 |