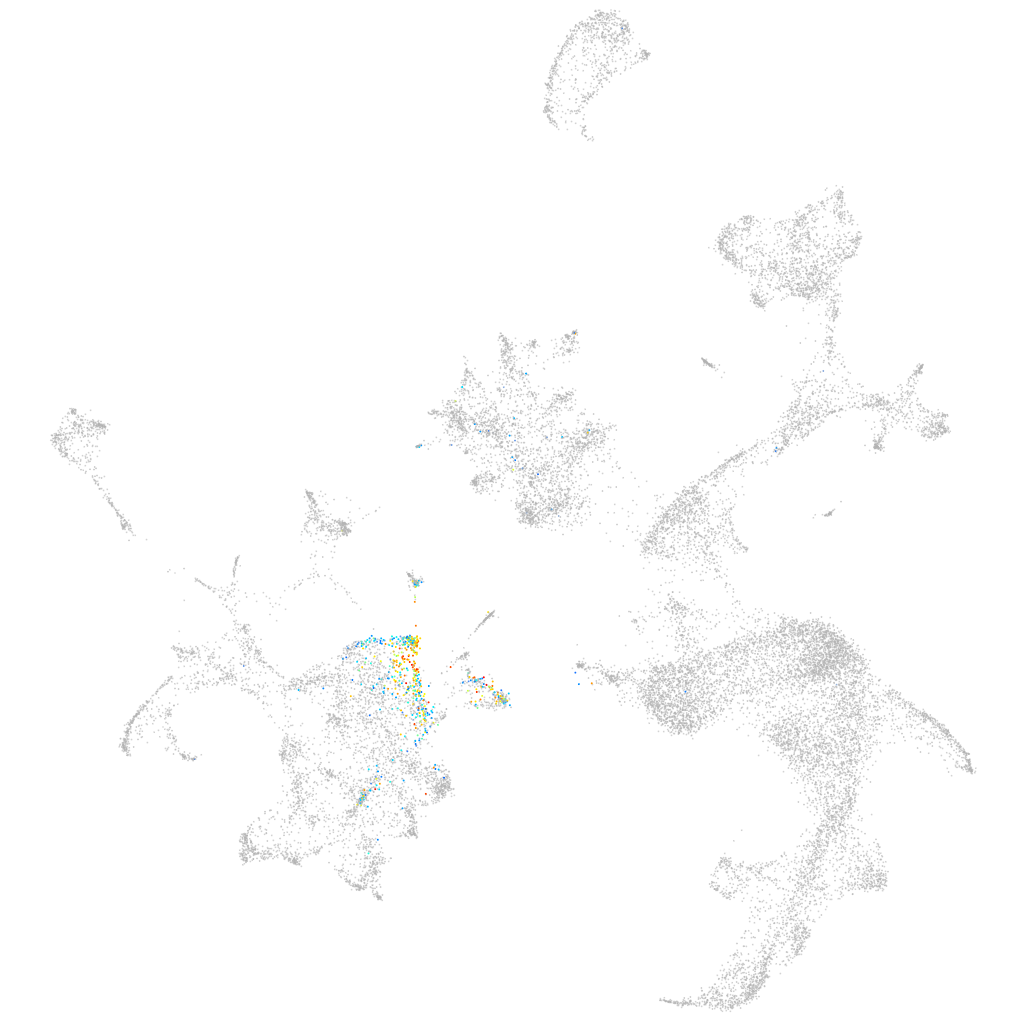
lymphatic vessel endothelial hyaluronic receptor 1a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lyve1b | 0.393 | hbae1.1 | -0.136 |
| gpr182 | 0.361 | hbae1.3 | -0.130 |
| ctgfa | 0.357 | hbae3 | -0.126 |
| stab1 | 0.353 | hbbe2 | -0.126 |
| cpn1 | 0.345 | hbbe1.3 | -0.122 |
| mrc1a | 0.339 | hbbe1.1 | -0.122 |
| plvapb | 0.321 | cahz | -0.120 |
| cldn11a | 0.320 | prdx2 | -0.114 |
| dab2 | 0.314 | hemgn | -0.113 |
| si:dkey-28n18.9 | 0.309 | hbbe1.2 | -0.113 |
| adma | 0.304 | blvrb | -0.111 |
| XLOC-016094 | 0.295 | alas2 | -0.106 |
| si:ch211-145b13.6 | 0.293 | tspo | -0.102 |
| loxl1 | 0.291 | gpx1a | -0.102 |
| sele | 0.289 | slc4a1a | -0.100 |
| fam174b | 0.280 | nt5c2l1 | -0.098 |
| hyal2b | 0.280 | fth1a | -0.097 |
| stab2 | 0.279 | si:ch211-250g4.3 | -0.095 |
| c1qtnf6a | 0.278 | epb41b | -0.094 |
| igf2b | 0.277 | zgc:163057 | -0.094 |
| fabp11a | 0.276 | si:ch211-207c6.2 | -0.090 |
| rgcc | 0.276 | zgc:56095 | -0.090 |
| lgmn | 0.276 | nmt1b | -0.084 |
| lfng | 0.263 | plac8l1 | -0.083 |
| lamp2 | 0.260 | tmod4 | -0.082 |
| ramp2 | 0.258 | creg1 | -0.080 |
| tie1 | 0.256 | hbae5 | -0.077 |
| ms4a17a.11 | 0.253 | sptb | -0.074 |
| lifrb | 0.251 | tdh | -0.074 |
| ednraa | 0.248 | znfl2a | -0.073 |
| tspan4b | 0.246 | uros | -0.073 |
| si:ch211-33e4.2 | 0.246 | hbbe3 | -0.073 |
| tgfbr2b | 0.239 | rfesd | -0.073 |
| gas6 | 0.239 | tfr1a | -0.073 |
| ap1b1 | 0.238 | tmem14ca | -0.071 |