hyaluronidase 2b
ZFIN 
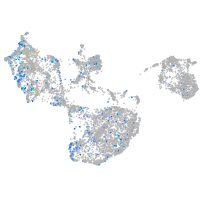
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152c2.3 | 0.182 | ckbb | -0.083 |
| hspb1 | 0.181 | gpm6aa | -0.082 |
| sp5l | 0.166 | stmn1b | -0.082 |
| nr6a1a | 0.163 | cyt1 | -0.079 |
| apela | 0.155 | krt4 | -0.079 |
| akap12b | 0.145 | tuba1c | -0.078 |
| crabp2b | 0.144 | rtn1a | -0.077 |
| npm1a | 0.139 | rnasekb | -0.075 |
| alcamb | 0.138 | atp6v0cb | -0.074 |
| fbl | 0.138 | hbbe1.3 | -0.072 |
| hoxb1b | 0.138 | cyt1l | -0.071 |
| chrd | 0.137 | icn | -0.071 |
| nop58 | 0.136 | pvalb1 | -0.071 |
| cdx4 | 0.135 | gpm6ab | -0.070 |
| dkc1 | 0.135 | hbae3 | -0.070 |
| nop56 | 0.132 | icn2 | -0.070 |
| pou5f3 | 0.132 | gng3 | -0.069 |
| ncl | 0.131 | krtt1c19e | -0.069 |
| nop2 | 0.131 | pvalb2 | -0.069 |
| stm | 0.131 | si:ch211-195b11.3 | -0.069 |
| pkdccb | 0.130 | sncb | -0.069 |
| msna | 0.128 | vamp2 | -0.067 |
| ebna1bp2 | 0.126 | gapdhs | -0.066 |
| zic2b | 0.126 | hbae1.1 | -0.066 |
| bms1 | 0.125 | elavl3 | -0.065 |
| BX927258.1 | 0.125 | anxa1a | -0.063 |
| cx43.4 | 0.125 | ccni | -0.063 |
| gnl3 | 0.125 | hbbe1.1 | -0.062 |
| zmp:0000000624 | 0.125 | atpv0e2 | -0.061 |
| her3 | 0.124 | agr1 | -0.060 |
| si:dkey-66i24.9 | 0.122 | anxa1c | -0.060 |
| gar1 | 0.121 | atp5if1b | -0.060 |
| lig1 | 0.121 | pycard | -0.060 |
| hnrnpa1b | 0.120 | krt5 | -0.059 |
| polr3gla | 0.120 | tcnbb | -0.059 |