hyaluronidase 2b
ZFIN 
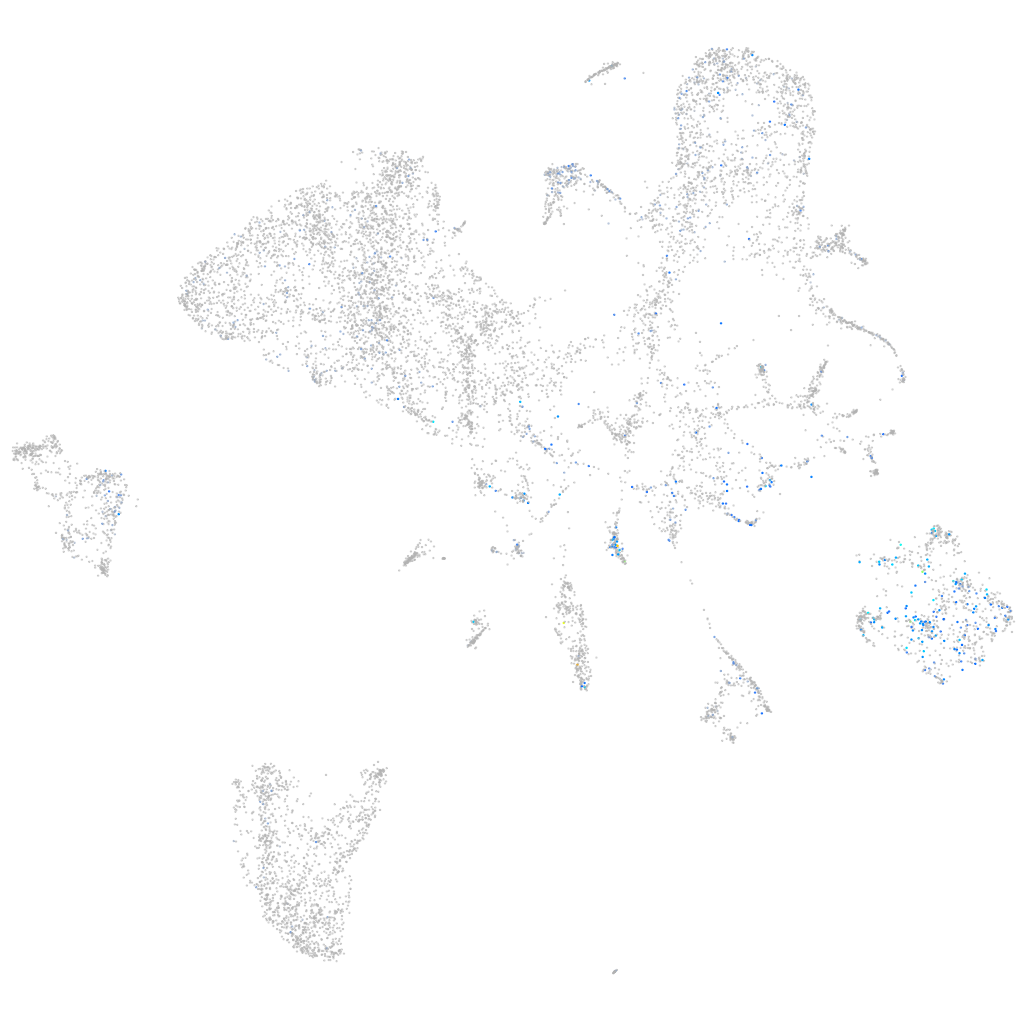
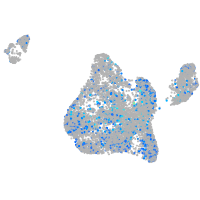
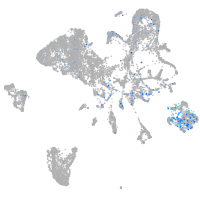
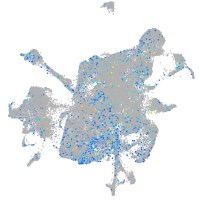
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh6 | 0.236 | rps10 | -0.186 |
| akap12b | 0.233 | rpl37 | -0.182 |
| si:ch211-152c2.3 | 0.226 | zgc:114188 | -0.164 |
| hspb1 | 0.226 | ahcy | -0.149 |
| asph | 0.214 | rps17 | -0.147 |
| marcksb | 0.205 | nme2b.1 | -0.139 |
| smo | 0.203 | gamt | -0.125 |
| smc1al | 0.200 | zgc:92744 | -0.124 |
| nr6a1a | 0.199 | nupr1b | -0.122 |
| NC-002333.4 | 0.199 | fabp3 | -0.122 |
| marcksl1b | 0.199 | eef1da | -0.120 |
| cx43.4 | 0.197 | gapdh | -0.117 |
| hnrnpa1a | 0.195 | bhmt | -0.110 |
| nucks1a | 0.195 | atp5l | -0.109 |
| ilf3b | 0.195 | zgc:158463 | -0.107 |
| tp53inp2 | 0.194 | gatm | -0.107 |
| hnrnpa1b | 0.191 | aqp12 | -0.107 |
| pltp | 0.188 | eno3 | -0.107 |
| acin1a | 0.188 | mat1a | -0.105 |
| hnrnpm | 0.188 | agxtb | -0.103 |
| stm | 0.187 | atp5if1b | -0.103 |
| hnrnpub | 0.185 | suclg1 | -0.098 |
| smarca4a | 0.185 | sod1 | -0.098 |
| qkia | 0.183 | apoa1b | -0.098 |
| syncrip | 0.182 | atp5fa1 | -0.098 |
| cxcr4a | 0.181 | grhprb | -0.097 |
| lmo4a | 0.181 | atp5mc3b | -0.096 |
| snrnp70 | 0.179 | aldh6a1 | -0.096 |
| seta | 0.178 | aldh7a1 | -0.096 |
| hp1bp3 | 0.178 | apoa2 | -0.095 |
| khdrbs1a | 0.177 | cox6a1 | -0.094 |
| top1l | 0.177 | gstt1a | -0.094 |
| ewsr1a | 0.176 | pklr | -0.093 |
| ppig | 0.174 | agxta | -0.093 |
| si:ch73-281n10.2 | 0.174 | COX7A2 (1 of many) | -0.093 |