Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 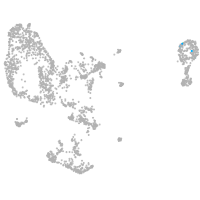 |
neural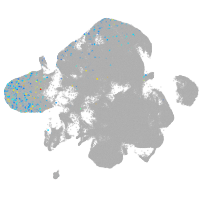 |
spinal cord / glia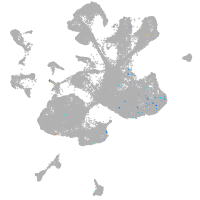 |
eye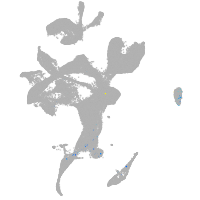 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mir457b | 0.602 | mt-co2 | -0.253 |
| gtse1 | 0.535 | dohh | -0.249 |
| dbpa | 0.527 | prpf3 | -0.222 |
| abhd11 | 0.511 | oaz1a | -0.216 |
| tut4 | 0.507 | pfn2l | -0.204 |
| aldh16a1 | 0.505 | atp5po | -0.201 |
| si:dkey-30f3.2 | 0.501 | dnaja2a | -0.199 |
| tmem136a | 0.497 | pomp | -0.194 |
| slc1a4 | 0.492 | ddx39ab | -0.194 |
| LOC110439496 | 0.465 | dr1 | -0.192 |
| CR339054.1 | 0.465 | cct6a | -0.188 |
| LOC110439797 | 0.465 | rfc5 | -0.179 |
| si:ch73-45o6.2 | 0.465 | ttf1 | -0.178 |
| sgk3 | 0.459 | npat | -0.176 |
| BX908770.1 | 0.458 | pabpn1 | -0.176 |
| CU655844.1 | 0.458 | wtap | -0.175 |
| pkd1l2b | 0.458 | rnf220a | -0.174 |
| znf1064 | 0.458 | numa1 | -0.173 |
| lhx8a | 0.444 | ndufa8 | -0.173 |
| vcpip1 | 0.444 | smarcad1a | -0.173 |
| BX511021.1 | 0.437 | prmt1 | -0.173 |
| chst2b | 0.436 | chmp2bb | -0.173 |
| BX914200.1 | 0.434 | ddx6 | -0.172 |
| angpt1 | 0.434 | morf4l1 | -0.170 |
| CR381647.1 | 0.434 | naa15a | -0.170 |
| CU137680.1 | 0.434 | atrx | -0.170 |
| dolk | 0.434 | upf3b | -0.169 |
| lgals3bpb | 0.434 | org | -0.169 |
| LOC103909508 | 0.434 | txndc9 | -0.169 |
| LOC108183703 | 0.434 | ufc1 | -0.168 |
| LOC110439352 | 0.434 | sox19b | -0.168 |
| nos1 | 0.434 | atp1a1a.1 | -0.167 |
| pip5kl1 | 0.434 | sod2 | -0.166 |
| rnf151 | 0.434 | pabpn1l | -0.165 |
| six1a | 0.434 | cript | -0.164 |