UPF3B regulator of nonsense mediated mRNA decay
ZFIN 
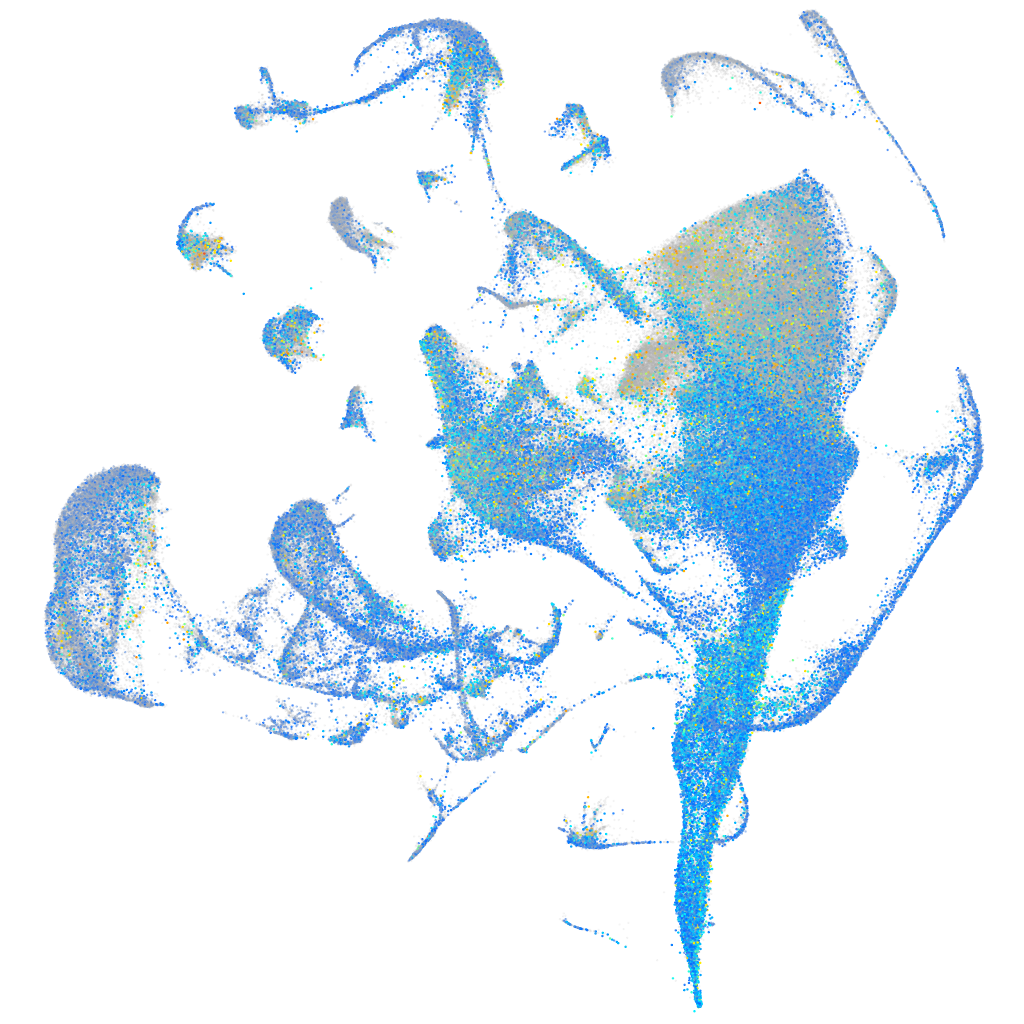
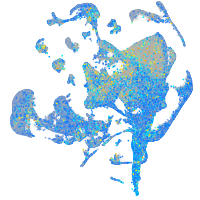
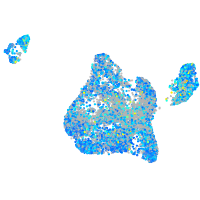
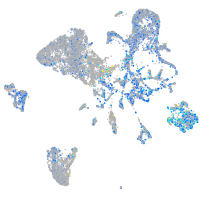
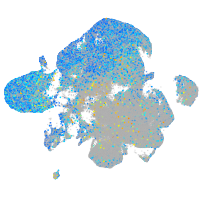
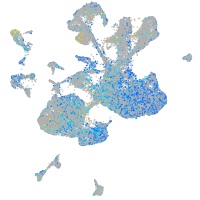
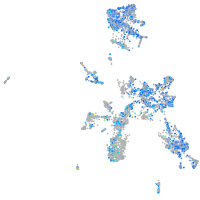
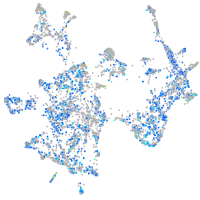
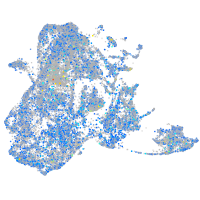
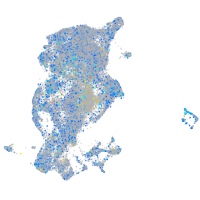
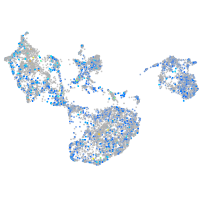
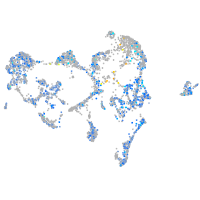
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nop58 | 0.178 | gng3 | -0.117 |
| lig1 | 0.174 | stmn1b | -0.116 |
| dkc1 | 0.173 | sncb | -0.112 |
| banf1 | 0.170 | atp6v0cb | -0.108 |
| mki67 | 0.170 | rtn1a | -0.108 |
| fbl | 0.168 | rtn1b | -0.102 |
| hnrnpa1b | 0.168 | elavl3 | -0.100 |
| hspb1 | 0.167 | ywhag2 | -0.100 |
| nop56 | 0.167 | elavl4 | -0.099 |
| anp32b | 0.166 | gpm6ab | -0.098 |
| chaf1a | 0.165 | vamp2 | -0.098 |
| ppig | 0.162 | ckbb | -0.097 |
| pou5f3 | 0.161 | rnasekb | -0.097 |
| zgc:56699 | 0.161 | stmn2a | -0.096 |
| npm1a | 0.160 | stx1b | -0.096 |
| akap12b | 0.159 | stxbp1a | -0.096 |
| cx43.4 | 0.159 | fez1 | -0.094 |
| s100a1 | 0.159 | atp6v1e1b | -0.093 |
| ccnd1 | 0.158 | mllt11 | -0.093 |
| ncl | 0.158 | gap43 | -0.092 |
| stm | 0.158 | gapdhs | -0.092 |
| zgc:110425 | 0.155 | pvalb1 | -0.092 |
| tpx2 | 0.154 | zgc:65894 | -0.092 |
| ccna2 | 0.153 | calm1a | -0.092 |
| cd2bp2 | 0.153 | snap25a | -0.090 |
| msna | 0.153 | atp6v1g1 | -0.089 |
| si:ch211-152c2.3 | 0.153 | gpm6aa | -0.089 |
| ccnb1 | 0.152 | pvalb2 | -0.089 |
| ppm1g | 0.152 | gng2 | -0.088 |
| dek | 0.151 | tuba1c | -0.088 |
| fbxo5 | 0.151 | tmsb2 | -0.087 |
| nasp | 0.151 | tmsb | -0.086 |
| ssrp1a | 0.151 | atpv0e2 | -0.084 |
| eif2s1a | 0.150 | olfm1b | -0.084 |
| gar1 | 0.150 | csdc2a | -0.083 |