Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia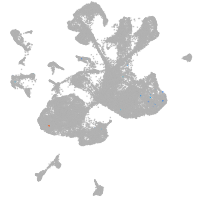 |
eye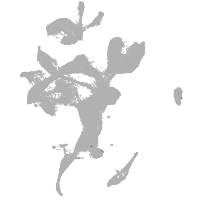 |
taste / olfactory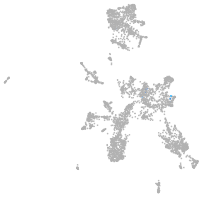 |
otic / lateral line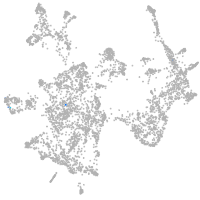 |
epidermis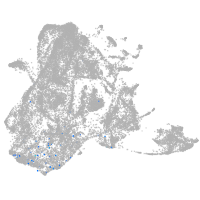 |
periderm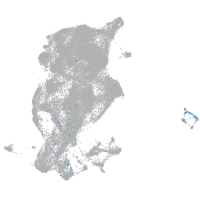 |
fin |
ionocytes / mucous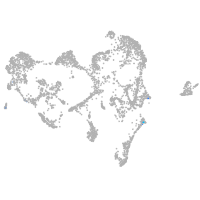 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR792441.1 | 0.149 | ckbb | -0.033 |
| hgfb | 0.147 | pvalb1 | -0.032 |
| LOC103910333 | 0.131 | gpm6aa | -0.032 |
| cacna1eb | 0.128 | actc1b | -0.028 |
| XLOC-028453 | 0.112 | tmsb4x | -0.028 |
| mpp7b | 0.098 | marcksl1b | -0.027 |
| ano8a | 0.088 | hbbe1.3 | -0.027 |
| hspb1 | 0.086 | fabp7a | -0.026 |
| adgre10 | 0.081 | CR383676.1 | -0.026 |
| LOC108182424 | 0.080 | CU467822.1 | -0.026 |
| BX927258.1 | 0.079 | hbae1.1 | -0.025 |
| XLOC-039121 | 0.075 | tmsb | -0.025 |
| cdx4 | 0.073 | pvalb2 | -0.025 |
| si:ch211-218h8.3 | 0.070 | fabp3 | -0.024 |
| XLOC-032526 | 0.068 | mylpfa | -0.024 |
| XLOC-034936 | 0.067 | CU634008.1 | -0.024 |
| XLOC-001603 | 0.065 | celf2 | -0.022 |
| LO018629.1 | 0.064 | h1f0 | -0.022 |
| apela | 0.062 | zgc:165461 | -0.021 |
| zgc:153151 | 0.062 | hbbe1.1 | -0.021 |
| CT573157.1 | 0.060 | rtn1a | -0.020 |
| si:ch211-132b12.1 | 0.057 | marcksl1a | -0.020 |
| gig2g | 0.056 | hbae3 | -0.020 |
| LOC110439343 | 0.053 | krt4 | -0.019 |
| LOC100332428 | 0.053 | gapdhs | -0.019 |
| znfl2a | 0.052 | gpm6ab | -0.019 |
| XLOC-018429 | 0.051 | tnni2a.4 | -0.019 |
| si:dkey-199f5.7 | 0.051 | atp1a1b | -0.018 |
| nradd | 0.051 | ppdpfb | -0.018 |
| sall4 | 0.051 | ost4 | -0.018 |
| noto | 0.051 | vat1 | -0.018 |
| il17d | 0.051 | sncb | -0.018 |
| ltbp1 | 0.050 | mdh1aa | -0.018 |
| BX005254.3 | 0.049 | rnasekb | -0.018 |
| admp | 0.049 | tnrc6c2 | -0.018 |




