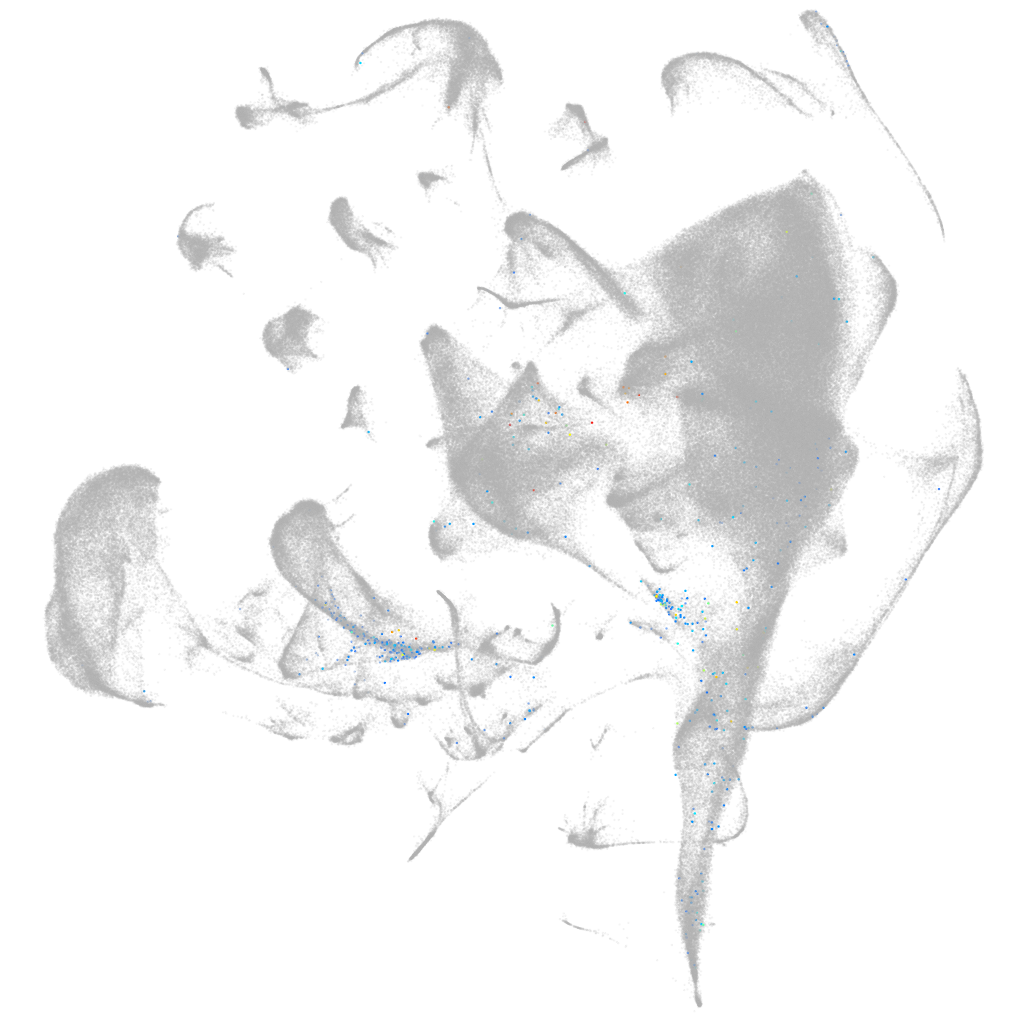
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm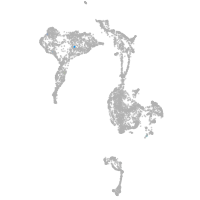 |
muscle 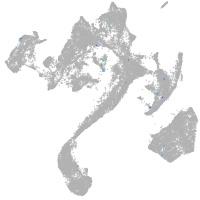 |
mural cells / non-skeletal muscle  |
mesenchyme 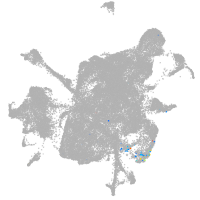 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis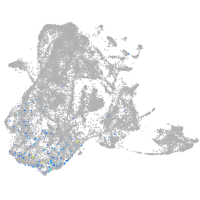 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fbn2a.1 | 0.118 | tuba1c | -0.021 |
| pth2r | 0.081 | pvalb1 | -0.020 |
| XLOC-028299 | 0.078 | pvalb2 | -0.020 |
| BX469925.3 | 0.073 | ckbb | -0.019 |
| fermt1 | 0.067 | gpm6aa | -0.019 |
| sdr16c5a | 0.064 | nova2 | -0.019 |
| LOC108180265 | 0.062 | elavl3 | -0.017 |
| si:dkey-12j5.1 | 0.062 | stmn1b | -0.017 |
| capn8 | 0.060 | hbbe1.3 | -0.016 |
| zgc:171775 | 0.060 | rnasekb | -0.016 |
| XLOC-000342 | 0.058 | rtn1a | -0.016 |
| pdlim1 | 0.055 | tmsb | -0.016 |
| cdh13 | 0.055 | actc1b | -0.015 |
| mir205 | 0.053 | gpm6ab | -0.015 |
| apoeb | 0.052 | tuba1a | -0.015 |
| bcl3 | 0.052 | cspg5a | -0.014 |
| mmp30 | 0.052 | fabp7a | -0.014 |
| cldni | 0.051 | gapdhs | -0.014 |
| CU137680.1 | 0.051 | gng3 | -0.014 |
| fn1a | 0.051 | hbae3 | -0.014 |
| tagln2 | 0.051 | ahcy | -0.013 |
| arhgef25b | 0.050 | atp6v0cb | -0.013 |
| cx44.2 | 0.049 | CU467822.1 | -0.013 |
| gpr31 | 0.049 | epb41a | -0.013 |
| myh9a | 0.049 | fabp3 | -0.013 |
| nid2a | 0.049 | hbae1.1 | -0.013 |
| BX005396.3 | 0.048 | mdkb | -0.013 |
| emilin2b | 0.048 | mylpfa | -0.013 |
| cdh1 | 0.047 | prdx2 | -0.013 |
| lama5 | 0.047 | si:ch211-137a8.4 | -0.013 |
| nuak1a | 0.047 | sncb | -0.013 |
| cnn2 | 0.046 | cadm3 | -0.012 |
| tp63 | 0.046 | elavl4 | -0.012 |
| XLOC-005272 | 0.046 | fez1 | -0.012 |
| XLOC-010030 | 0.046 | gnao1a | -0.012 |