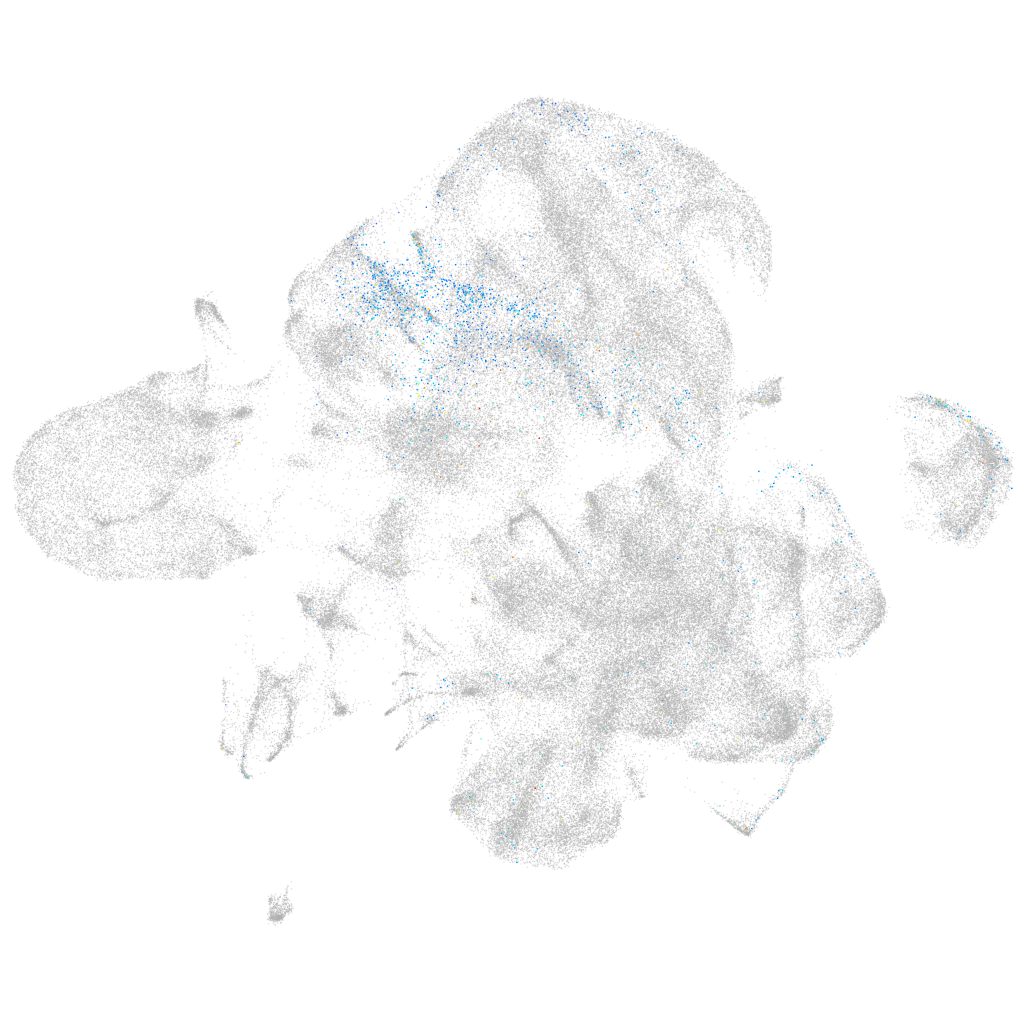
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural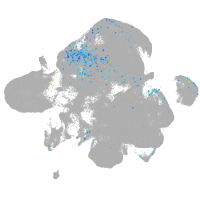 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line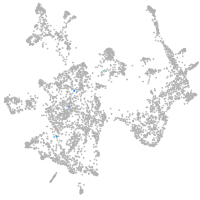 |
epidermis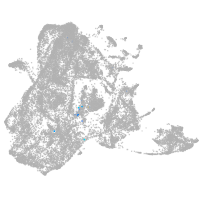 |
periderm |
fin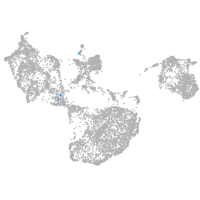 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-69b22.5 | 0.190 | marcksl1b | -0.049 |
| fabp11a | 0.145 | sncb | -0.041 |
| zic6 | 0.139 | rtn1b | -0.041 |
| cep112 | 0.111 | gpm6ab | -0.039 |
| zic2a | 0.108 | atp6v0cb | -0.038 |
| clybl | 0.104 | gng3 | -0.038 |
| tnfrsf19 | 0.102 | elavl4 | -0.036 |
| mdka | 0.097 | ckbb | -0.035 |
| zic1 | 0.096 | stx1b | -0.035 |
| zic4 | 0.095 | vamp2 | -0.035 |
| zic3 | 0.091 | stmn2a | -0.034 |
| zic5 | 0.087 | stxbp1a | -0.034 |
| fzd10 | 0.078 | h2afx1 | -0.034 |
| BX511142.1 | 0.072 | ywhag2 | -0.034 |
| si:ch211-156b7.4 | 0.068 | snap25a | -0.033 |
| stmn1a | 0.067 | atp6v1e1b | -0.033 |
| tenm3 | 0.067 | tmem59l | -0.033 |
| smoc2 | 0.066 | gap43 | -0.032 |
| ahcy | 0.066 | si:dkeyp-75h12.5 | -0.032 |
| LOC101882140 | 0.065 | zc4h2 | -0.032 |
| msi1 | 0.065 | map1aa | -0.031 |
| her15.1 | 0.064 | gng2 | -0.031 |
| her2 | 0.064 | nsg2 | -0.031 |
| trabd2a | 0.063 | nova1 | -0.031 |
| si:ch211-66e2.5 | 0.062 | zgc:65894 | -0.031 |
| wls | 0.062 | rtn1a | -0.031 |
| sema3bl | 0.061 | tmsb2 | -0.030 |
| BX469902.1 | 0.061 | hoxb3a | -0.030 |
| atp1a1b | 0.060 | nrxn1a | -0.030 |
| zgc:165461 | 0.060 | aplp1 | -0.030 |
| cxcl12a | 0.060 | olfm1b | -0.030 |
| tuba8l | 0.059 | mllt11 | -0.029 |
| lrig1 | 0.059 | dpysl3 | -0.029 |
| pdgfaa | 0.057 | id4 | -0.029 |
| podxl | 0.056 | gabrb4 | -0.029 |