Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 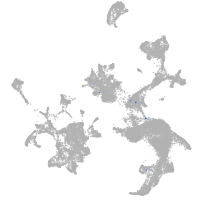 |
pronephros 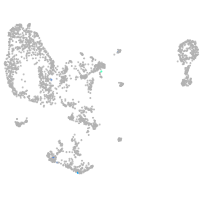 |
neural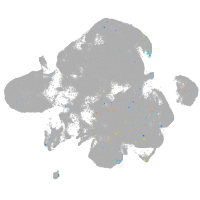 |
spinal cord / glia |
eye |
taste / olfactory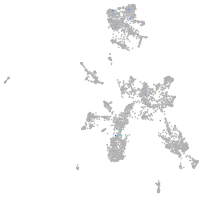 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fam19a1a | 0.904 | khdrbs1a | -0.410 |
| atp6ap1a | 0.903 | hmgb2b | -0.302 |
| hbbe1.2 | 0.896 | seta | -0.287 |
| cers1 | 0.888 | stmn1a | -0.286 |
| xirp2a | 0.874 | h2afva | -0.285 |
| adrb2b | 0.859 | mki67 | -0.270 |
| CABZ01026681.1 | 0.859 | snrpb | -0.268 |
| CU639468.1 | 0.859 | dnajc1 | -0.263 |
| cxcr3.2 | 0.859 | rpl12 | -0.259 |
| dvl1a | 0.859 | si:ch211-288g17.3 | -0.258 |
| ecrg4b | 0.859 | ranbp1 | -0.258 |
| elfn1a | 0.859 | hnrnpaba | -0.254 |
| gpr183b | 0.859 | h1m | -0.252 |
| lrtm2a | 0.859 | ppig | -0.247 |
| nkx2.3 | 0.859 | ccna2 | -0.241 |
| nlgn3a | 0.859 | eif2s2 | -0.240 |
| ntng2b | 0.859 | khdrbs1b | -0.240 |
| prmt8b | 0.859 | zmat2 | -0.239 |
| prss12 | 0.859 | hmgb2a | -0.235 |
| prune2 | 0.859 | cx43.4 | -0.233 |
| rgs16 | 0.859 | chaf1a | -0.233 |
| scg2b | 0.859 | snrpb2 | -0.232 |
| sema5a | 0.859 | snu13b | -0.230 |
| si:ch211-270n8.1 | 0.859 | hnrnpa1a | -0.230 |
| si:dkey-253d23.3 | 0.859 | zgc:56699 | -0.229 |
| syt12 | 0.859 | fth1a | -0.228 |
| tmsb | 0.859 | baz1b | -0.227 |
| XLOC-043619 | 0.859 | hnrnpa1b | -0.226 |
| LOC103911479 | 0.859 | hspb1 | -0.225 |
| si:ch211-51a6.2 | 0.859 | hnrnph1l | -0.222 |
| hapln1b | 0.851 | hdac1 | -0.221 |
| alkal2a | 0.850 | hnrnpm | -0.220 |
| btk | 0.845 | nasp | -0.220 |
| gpx3 | 0.839 | odc1 | -0.219 |
| mat1a | 0.839 | matr3l1.1 | -0.218 |