5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
ZFIN 
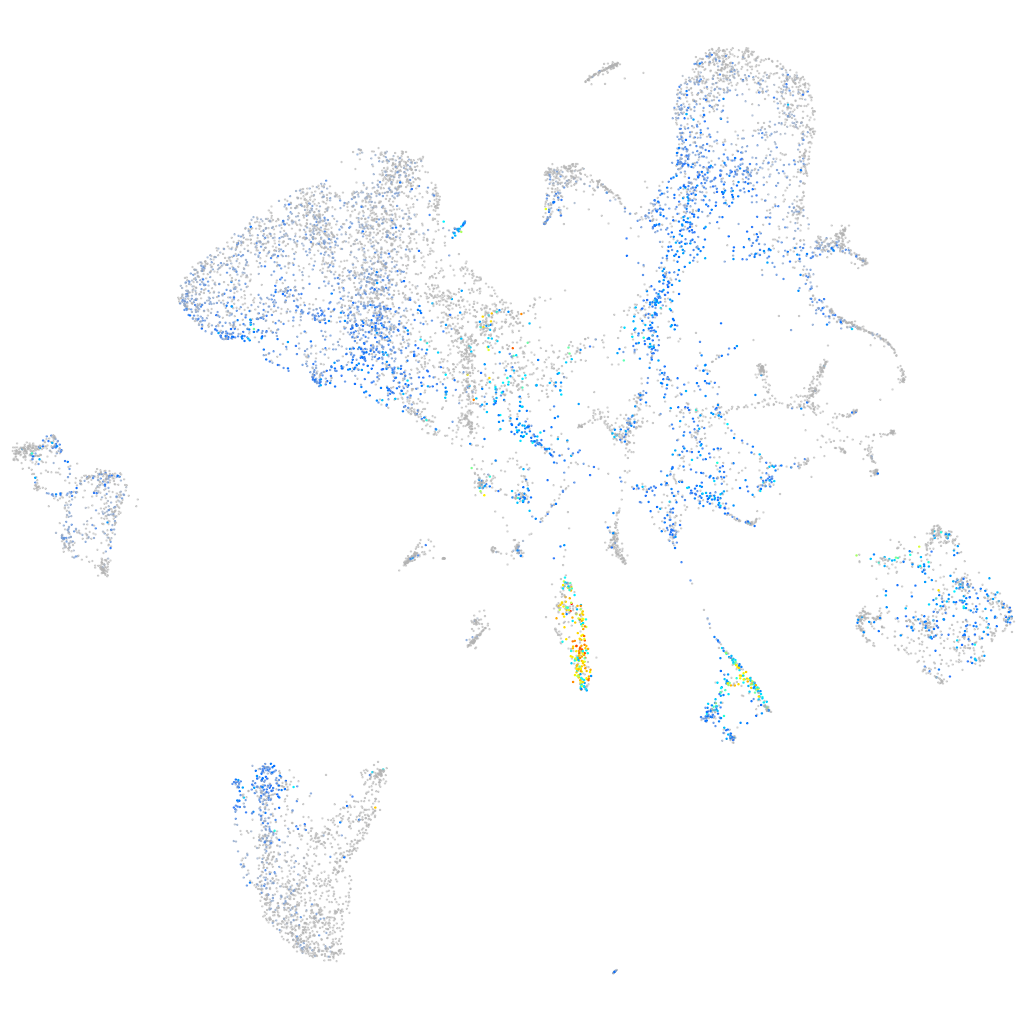
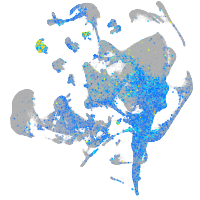
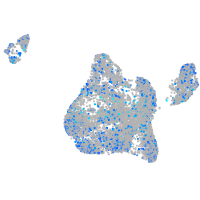
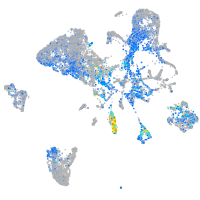
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paics | 0.466 | pnp4b | -0.177 |
| pvalb9 | 0.405 | cyp3a65 | -0.167 |
| gart | 0.397 | dedd1 | -0.149 |
| prps1a | 0.378 | cyp2n13 | -0.148 |
| myl9b | 0.369 | chac1 | -0.146 |
| si:dkey-126g1.9 | 0.356 | zgc:136461 | -0.140 |
| zgc:136410 | 0.352 | gda | -0.135 |
| acp5a | 0.348 | prss1 | -0.133 |
| cst14b.1 | 0.347 | ela2l | -0.133 |
| zgc:114181 | 0.332 | ctrb1 | -0.131 |
| tktb | 0.328 | cyp2ad2 | -0.131 |
| si:dkey-31g6.6 | 0.327 | prss59.2 | -0.130 |
| myl6 | 0.324 | cldnh | -0.130 |
| ldhba | 0.322 | prss59.1 | -0.130 |
| dao.1 | 0.313 | CELA1 (1 of many) | -0.129 |
| ctss2.1 | 0.308 | amy2a | -0.129 |
| rrm2b | 0.294 | ela2 | -0.128 |
| rbp5 | 0.289 | ela3l | -0.127 |
| fahd2a | 0.287 | c6ast4 | -0.127 |
| mthfd1l | 0.285 | spink4 | -0.126 |
| adsl | 0.283 | sycn.2 | -0.126 |
| mia | 0.280 | zgc:112160 | -0.126 |
| slc13a4 | 0.276 | COX3 | -0.125 |
| psph | 0.271 | zgc:92137 | -0.124 |
| gskip | 0.265 | eif4ebp3 | -0.123 |
| nt5c3a | 0.262 | tdo2b | -0.122 |
| txn | 0.261 | pdk2a | -0.121 |
| apoa1a | 0.259 | pla2g1b | -0.121 |
| cad | 0.255 | cpb1 | -0.119 |
| pfas | 0.255 | faxdc2 | -0.119 |
| dbi | 0.252 | slc2a9l2 | -0.119 |
| capn2a | 0.252 | ucp1 | -0.118 |
| ctsll | 0.250 | lpin1 | -0.118 |
| CABZ01048402.2 | 0.248 | cpa1 | -0.117 |
| fabp11a | 0.244 | ckba | -0.117 |