asparaginase homolog (S. cerevisiae)
ZFIN 
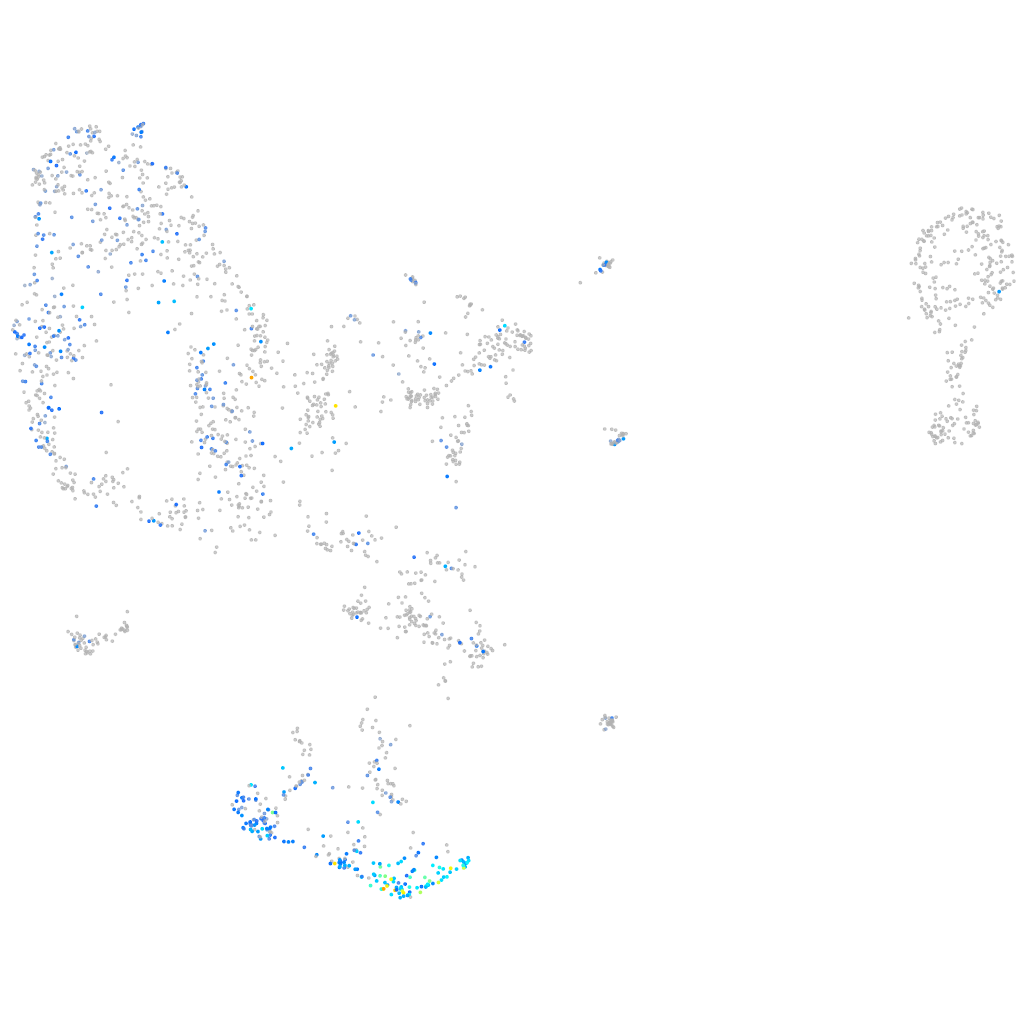
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glsb | 0.581 | si:ch73-1a9.3 | -0.234 |
| zgc:158423 | 0.572 | hnrnpabb | -0.221 |
| slc9a3.2 | 0.555 | si:ch73-281n10.2 | -0.211 |
| trim35-12 | 0.552 | eif5a2 | -0.210 |
| slc7a8a | 0.549 | marcksl1b | -0.208 |
| mal2 | 0.548 | marcksb | -0.204 |
| rhcgb | 0.532 | hspb1 | -0.200 |
| soul2 | 0.523 | nucks1a | -0.197 |
| si:dkey-36i7.3 | 0.516 | ptmab | -0.192 |
| etf1a | 0.506 | snrpf | -0.188 |
| ccl19a.1 | 0.506 | wu:fb97g03 | -0.188 |
| bsnd | 0.487 | snrpg | -0.186 |
| atp1a1a.3 | 0.487 | hnrnpaba | -0.183 |
| atp1a1a.5 | 0.485 | msna | -0.181 |
| phlda2 | 0.484 | ilf3b | -0.179 |
| slc12a10.3 | 0.478 | hmgn2 | -0.177 |
| sfxn3 | 0.469 | rps12 | -0.176 |
| tmem72 | 0.468 | h2afvb | -0.175 |
| clcnk | 0.460 | cx43.4 | -0.174 |
| ca4c | 0.460 | hmgb2b | -0.174 |
| ckmt1 | 0.457 | apoeb | -0.174 |
| degs2 | 0.454 | syncrip | -0.170 |
| glud1a | 0.450 | acy3.2 | -0.168 |
| tspan13a | 0.448 | setb | -0.165 |
| BX000438.2 | 0.444 | hmga1a | -0.165 |
| alas1 | 0.443 | snrnp70 | -0.164 |
| me3 | 0.442 | ptges3b | -0.164 |
| rnf183 | 0.435 | h3f3d | -0.164 |
| tmem176l.4 | 0.433 | akap12b | -0.164 |
| gadd45bb | 0.433 | nono | -0.163 |
| si:ch73-359m17.9 | 0.432 | si:ch211-222l21.1 | -0.163 |
| thbs1b | 0.427 | aqp8b | -0.162 |
| sorl1 | 0.426 | zgc:110425 | -0.159 |
| ctss2.2 | 0.417 | fbl | -0.158 |
| ppp1r1b | 0.413 | top1l | -0.158 |