apelin receptor early endogenous ligand
ZFIN 
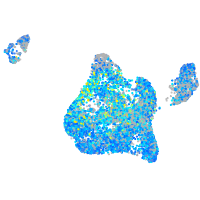
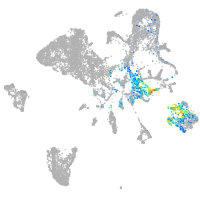
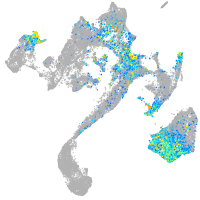
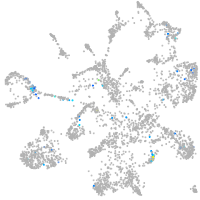
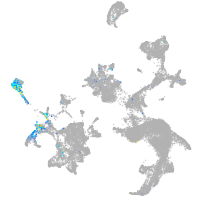
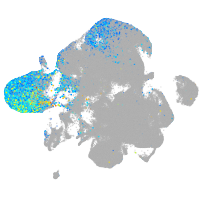
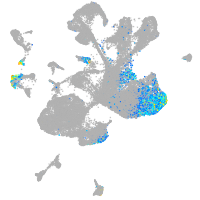
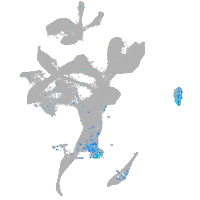
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdx4 | 0.553 | ahcy | -0.315 |
| cdh6 | 0.534 | rpl37 | -0.312 |
| pltp | 0.497 | rps10 | -0.299 |
| hspb1 | 0.482 | gapdh | -0.285 |
| cx43.4 | 0.449 | eno3 | -0.280 |
| fgfrl1b | 0.438 | nme2b.1 | -0.279 |
| hapln1b | 0.437 | sod1 | -0.276 |
| akap12b | 0.435 | eef1da | -0.275 |
| prickle1a | 0.428 | zgc:114188 | -0.275 |
| hoxc8a | 0.427 | rps17 | -0.271 |
| nradd | 0.423 | gamt | -0.269 |
| oc90 | 0.416 | atp5if1b | -0.268 |
| sept15 | 0.416 | suclg1 | -0.243 |
| qkia | 0.415 | gatm | -0.242 |
| marcksb | 0.414 | aldob | -0.242 |
| si:ch211-152c2.3 | 0.408 | nupr1b | -0.240 |
| tgif1 | 0.403 | bhmt | -0.235 |
| crtap | 0.401 | tpi1b | -0.230 |
| wnt5b | 0.398 | gstp1 | -0.229 |
| znfl2a | 0.398 | apoa1b | -0.227 |
| tpm4a | 0.395 | zgc:158463 | -0.226 |
| marcksl1b | 0.392 | dap | -0.225 |
| emid1 | 0.392 | gpx4a | -0.225 |
| nucks1a | 0.392 | fbp1b | -0.224 |
| fgf10b | 0.391 | mdh1aa | -0.222 |
| lypd6b | 0.388 | glud1b | -0.221 |
| prickle1b | 0.385 | sod2 | -0.220 |
| si:ch73-281n10.2 | 0.379 | mat1a | -0.219 |
| ndnf | 0.377 | atp5mc1 | -0.218 |
| serpinh1b | 0.373 | mt-atp6 | -0.216 |
| efna5a | 0.373 | apoa4b.1 | -0.213 |
| hoxc3a | 0.372 | aldh6a1 | -0.211 |
| id3 | 0.371 | fabp3 | -0.211 |
| syncrip | 0.371 | atp5pf | -0.210 |
| NC-002333.4 | 0.371 | scp2a | -0.210 |