adaptor related protein complex 3 subunit mu 2
ZFIN 
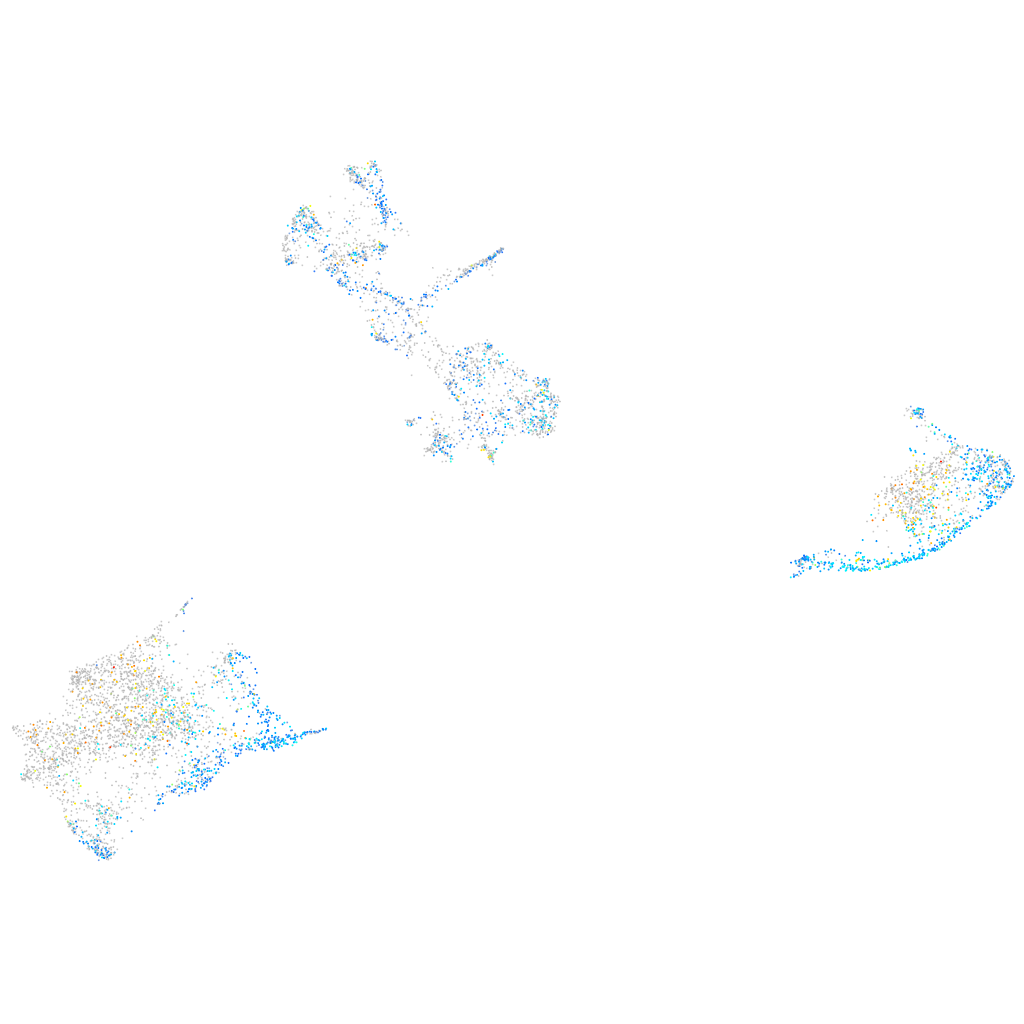
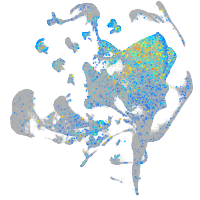
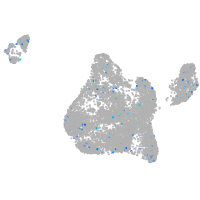
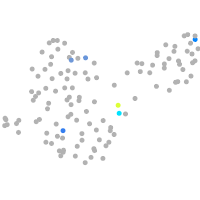
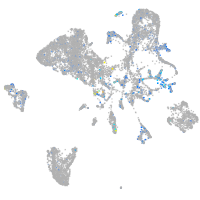
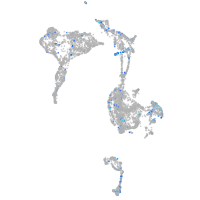
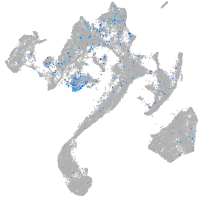
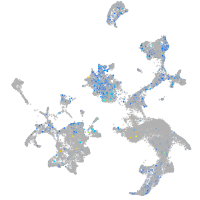
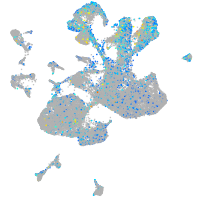
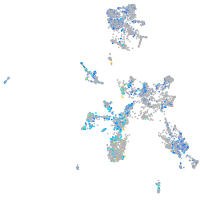
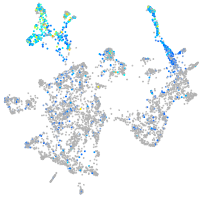
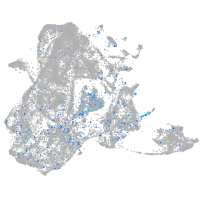
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmem243b | 0.275 | pvalb2 | -0.139 |
| mitfa | 0.250 | pvalb1 | -0.137 |
| atp6v1g1 | 0.247 | CABZ01021592.1 | -0.112 |
| tfap2e | 0.240 | actc1b | -0.110 |
| slc37a2 | 0.234 | si:dkey-251i10.2 | -0.108 |
| tyr | 0.229 | uraha | -0.107 |
| qdpra | 0.229 | mylpfa | -0.098 |
| msx1b | 0.227 | hbbe1.3 | -0.090 |
| atp6v1ba | 0.225 | mylz3 | -0.087 |
| tspan36 | 0.224 | mdh1aa | -0.086 |
| rab38 | 0.221 | aqp3a | -0.083 |
| rab32a | 0.221 | hbae3 | -0.082 |
| atp6v0ca | 0.217 | si:ch211-251b21.1 | -0.081 |
| rabl6b | 0.217 | apoa2 | -0.081 |
| pah | 0.217 | phyhd1 | -0.080 |
| xbp1 | 0.214 | myhz1.1 | -0.080 |
| slc45a2 | 0.213 | krt91 | -0.077 |
| atp6v1c1b | 0.212 | mdka | -0.076 |
| dct | 0.212 | hbbe2 | -0.075 |
| tmem243a | 0.212 | prss59.2 | -0.074 |
| slc7a5 | 0.211 | krt18b | -0.074 |
| naga | 0.210 | ptmaa | -0.071 |
| atp6ap2 | 0.209 | ndrg2 | -0.070 |
| atp6v0a2b | 0.207 | apoa1b | -0.069 |
| SPAG9 | 0.205 | ttn.1 | -0.067 |
| gpr143 | 0.205 | tnni2a.4 | -0.067 |
| tspan10 | 0.204 | tnnt3b | -0.066 |
| sypl2b | 0.203 | ckmb | -0.066 |
| pmela | 0.203 | defbl1 | -0.066 |
| slc3a2a | 0.202 | krt8 | -0.065 |
| atp6ap1b | 0.202 | tpma | -0.065 |
| rnaseka | 0.200 | apoda.1 | -0.065 |
| stx12 | 0.199 | hbbe1.1 | -0.065 |
| lamp1a | 0.199 | agmo | -0.064 |
| bace2 | 0.198 | ctrb1 | -0.063 |