adaptor related protein complex 3 subunit mu 2
ZFIN 
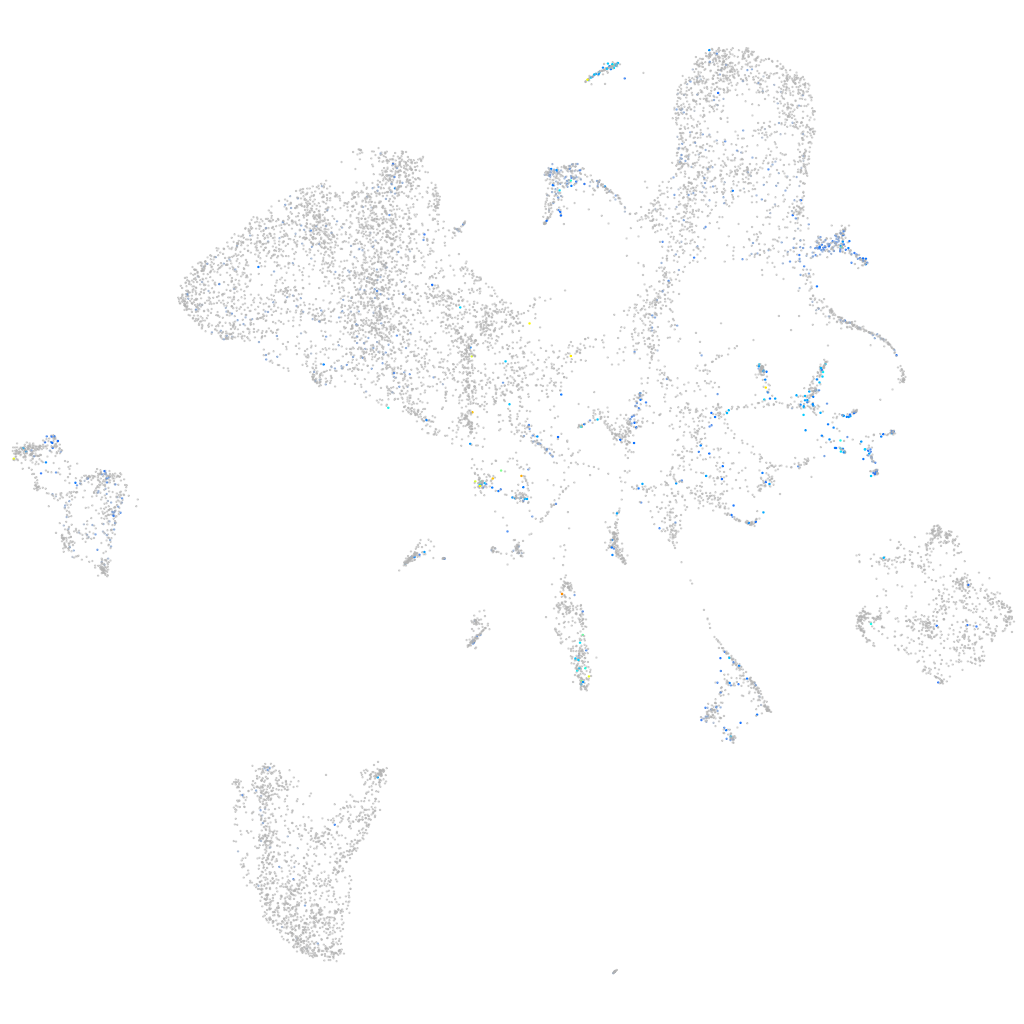
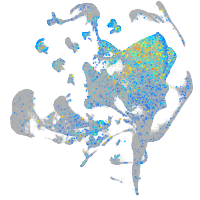
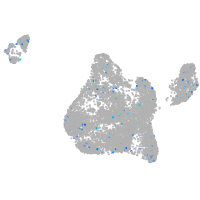
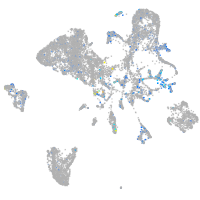
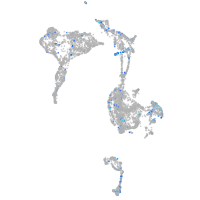
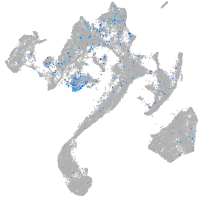
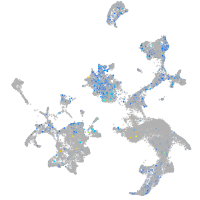
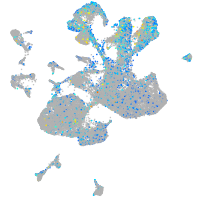
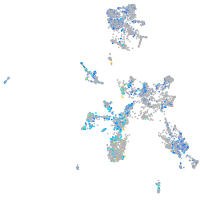
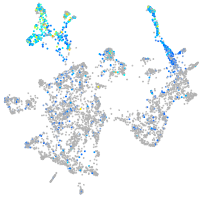
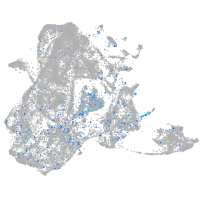
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.252 | gamt | -0.112 |
| neurod1 | 0.228 | gapdh | -0.111 |
| pax6b | 0.217 | gatm | -0.110 |
| fev | 0.215 | ahcy | -0.109 |
| mir375-2 | 0.212 | agxtb | -0.106 |
| zgc:101731 | 0.210 | mat1a | -0.106 |
| rnasekb | 0.207 | pnp4b | -0.104 |
| gpc1a | 0.206 | aqp12 | -0.103 |
| gapdhs | 0.204 | nupr1b | -0.098 |
| pcsk1nl | 0.203 | bhmt | -0.095 |
| calm1b | 0.201 | glud1b | -0.093 |
| insm1a | 0.201 | fbp1b | -0.091 |
| gdi1 | 0.200 | ttc36 | -0.090 |
| scgn | 0.198 | agxta | -0.089 |
| atp6ap2 | 0.198 | aldh6a1 | -0.088 |
| c2cd4a | 0.196 | fabp10a | -0.087 |
| egr4 | 0.195 | gcshb | -0.087 |
| insm1b | 0.190 | apoc1 | -0.086 |
| si:dkey-153k10.9 | 0.189 | cyp2n13 | -0.086 |
| si:ch1073-429i10.3.1 | 0.188 | scp2a | -0.085 |
| id4 | 0.187 | gc | -0.085 |
| hsbp1a | 0.185 | cyp4v8 | -0.085 |
| pygma | 0.185 | gstt1a | -0.085 |
| mir7a-1 | 0.184 | igfbp2a | -0.085 |
| elovl1a | 0.181 | uox | -0.084 |
| necap1 | 0.180 | aldob | -0.084 |
| jagn1a | 0.179 | apoa1b | -0.084 |
| kcnj11 | 0.178 | ttr | -0.084 |
| vat1 | 0.178 | ppdpfa | -0.084 |
| syt1a | 0.178 | si:dkey-16p21.8 | -0.084 |
| gnao1a | 0.177 | rbp2b | -0.083 |
| tox | 0.177 | si:dkey-86l18.10 | -0.083 |
| myt1b | 0.176 | cpn1 | -0.083 |
| lysmd2 | 0.175 | mgst1.2 | -0.083 |
| oaz1b | 0.174 | LOC110437731 | -0.082 |