ankyrin repeat and EF-hand domain containing 1a
ZFIN 
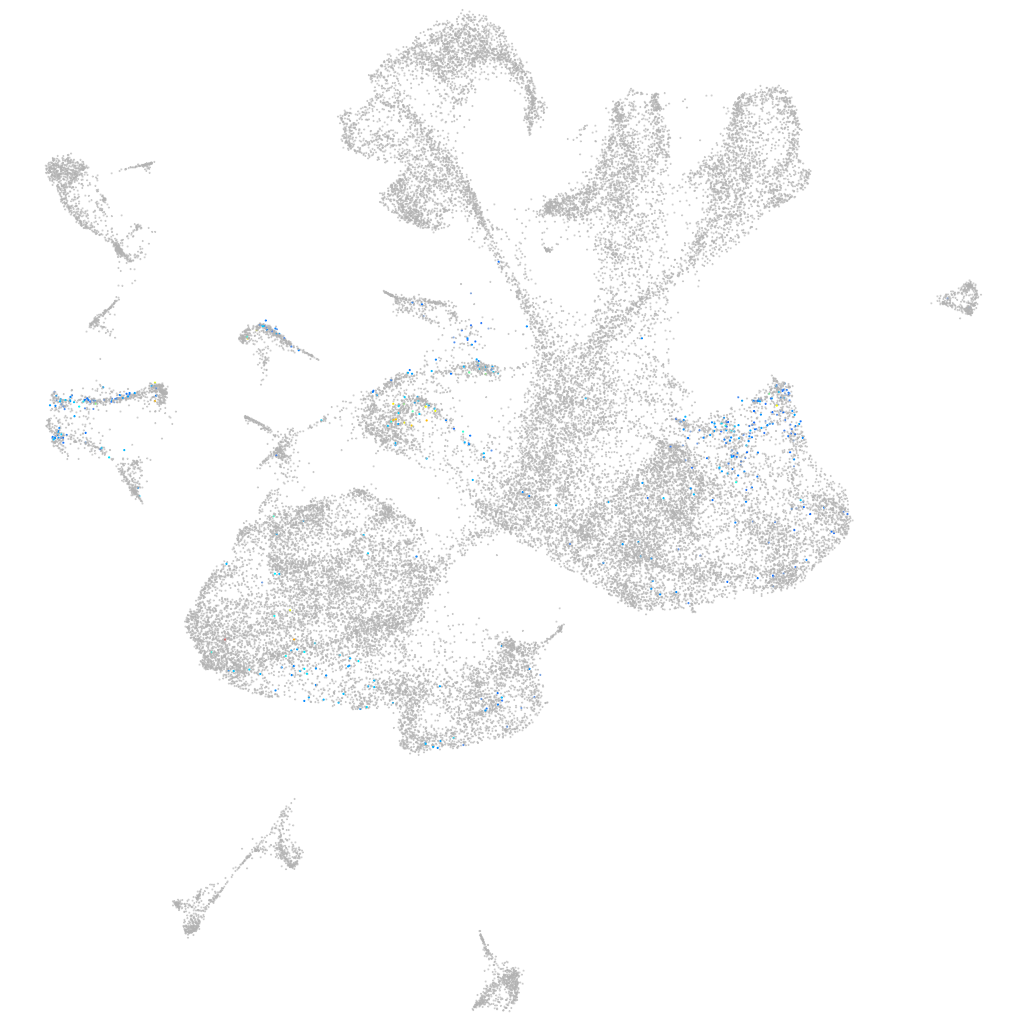
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm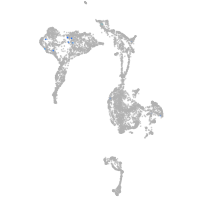 |
muscle 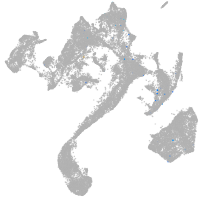 |
mural cells / non-skeletal muscle 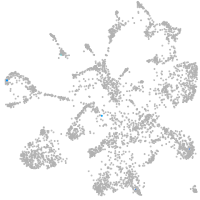 |
mesenchyme 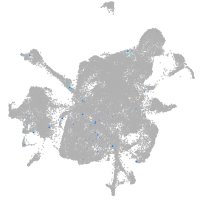 |
hematopoietic / vasculature  |
pronephros 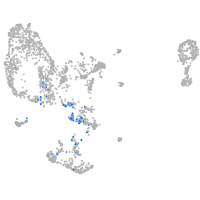 |
neural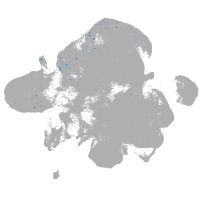 |
spinal cord / glia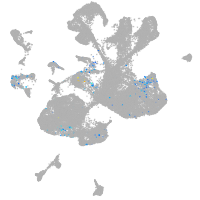 |
eye |
taste / olfactory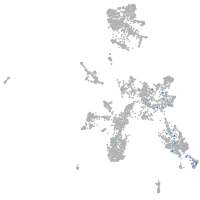 |
otic / lateral line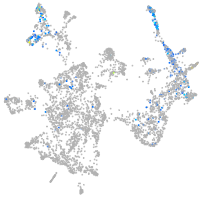 |
epidermis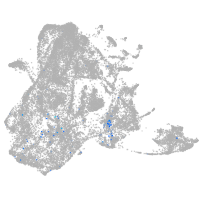 |
periderm |
fin |
ionocytes / mucous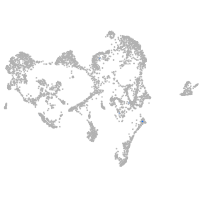 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| enkur | 0.168 | elavl3 | -0.059 |
| tbata | 0.164 | tmsb4x | -0.057 |
| daw1 | 0.162 | rtn1a | -0.055 |
| si:ch211-248e11.2 | 0.151 | stmn1b | -0.053 |
| smkr1 | 0.150 | tmsb | -0.050 |
| dnali1 | 0.149 | hmgb3a | -0.050 |
| efcab1 | 0.147 | myt1b | -0.049 |
| foxj1a | 0.143 | si:dkey-276j7.1 | -0.048 |
| catip | 0.143 | gpm6ab | -0.045 |
| si:dkey-148f10.4 | 0.143 | ywhah | -0.042 |
| spata18 | 0.141 | gng3 | -0.042 |
| si:ch211-163l21.7 | 0.140 | fabp3 | -0.042 |
| ccdc173 | 0.139 | tubb5 | -0.041 |
| pacrg | 0.137 | ptmaa | -0.040 |
| si:ch211-155m12.5 | 0.136 | vamp2 | -0.040 |
| zgc:55461 | 0.133 | stxbp1a | -0.039 |
| rab36 | 0.133 | stx1b | -0.039 |
| cfap157 | 0.131 | tmeff1b | -0.038 |
| ak7a | 0.131 | rtn1b | -0.037 |
| si:dkeyp-69c1.9 | 0.130 | sncb | -0.037 |
| krt18a.1 | 0.129 | atp6v0cb | -0.037 |
| lrrc6 | 0.126 | elavl4 | -0.036 |
| cfap126 | 0.126 | nsg2 | -0.036 |
| pih1d3 | 0.124 | ptmab | -0.036 |
| meig1 | 0.124 | myt1a | -0.036 |
| si:ch211-163l21.10 | 0.124 | syt2a | -0.036 |
| rsph4a | 0.123 | si:ch73-386h18.1 | -0.036 |
| cfap45 | 0.122 | snap25a | -0.036 |
| or128-8 | 0.122 | gng2 | -0.035 |
| ccdc114 | 0.122 | nova2 | -0.035 |
| si:ch73-265d7.2 | 0.121 | nova1 | -0.035 |
| si:dkey-42p14.3 | 0.121 | onecut2 | -0.035 |
| capsla | 0.121 | nkain1 | -0.035 |
| ttc25 | 0.121 | tmem59l | -0.035 |
| LOC103910812 | 0.120 | atcaya | -0.034 |