ankyrin repeat and EF-hand domain containing 1a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural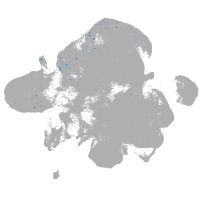 |
spinal cord / glia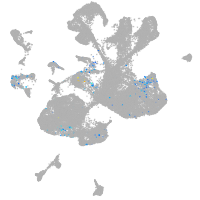 |
eye |
taste / olfactory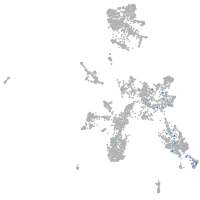 |
otic / lateral line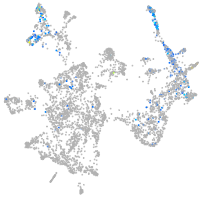 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rab38c | 0.884 | calm2a | -0.201 |
| thbs3b | 0.822 | hnrnpaba | -0.183 |
| ACKR2 | 0.765 | banf1 | -0.181 |
| adarb1b | 0.765 | marcksb | -0.163 |
| AL590150.1 | 0.765 | gar1 | -0.163 |
| AL929304.1 | 0.765 | actb2 | -0.163 |
| arhgef3 | 0.765 | zgc:56095 | -0.162 |
| bsna | 0.765 | zgc:110425 | -0.157 |
| BX323991.1 | 0.765 | eef1a1l1 | -0.156 |
| BX470229.2 | 0.765 | sec62 | -0.155 |
| BX510909.2 | 0.765 | CABZ01044053.1 | -0.155 |
| BX530017.2 | 0.765 | npc2 | -0.152 |
| BX530024.1 | 0.765 | hnrnpa0b | -0.149 |
| BX927308.1 | 0.765 | ddx61 | -0.148 |
| cd247l | 0.765 | zgc:173742 | -0.147 |
| cdc42se2 | 0.765 | brd7 | -0.147 |
| CR589875.1 | 0.765 | srfbp1 | -0.145 |
| crybb2 | 0.765 | ptp4a1 | -0.145 |
| diras1b | 0.765 | rhoaa | -0.144 |
| dkk1a | 0.765 | srsf5a | -0.143 |
| dock11 | 0.765 | hmga1a | -0.142 |
| fam151a | 0.765 | arf1 | -0.141 |
| FO834888.1 | 0.765 | ddx23 | -0.140 |
| gabbr1a | 0.765 | cbx1a | -0.139 |
| glra1 | 0.765 | thoc2 | -0.138 |
| gramd4a | 0.765 | cstf3 | -0.138 |
| helt | 0.765 | srsf2a | -0.137 |
| il7r | 0.765 | ybx1 | -0.137 |
| kcnk13b | 0.765 | akap12b | -0.136 |
| LOC100331003 | 0.765 | hnrnpc | -0.134 |
| LOC101884549 | 0.765 | ptmab | -0.133 |
| LOC101885174 | 0.765 | tpt1 | -0.132 |
| LOC110438236 | 0.765 | stmn1a | -0.132 |
| LOC110439812 | 0.765 | AL935174.5 | -0.131 |
| LOC110439944 | 0.765 | marcksl1b | -0.131 |




