adhesion G protein-coupled receptor L2a
ZFIN 
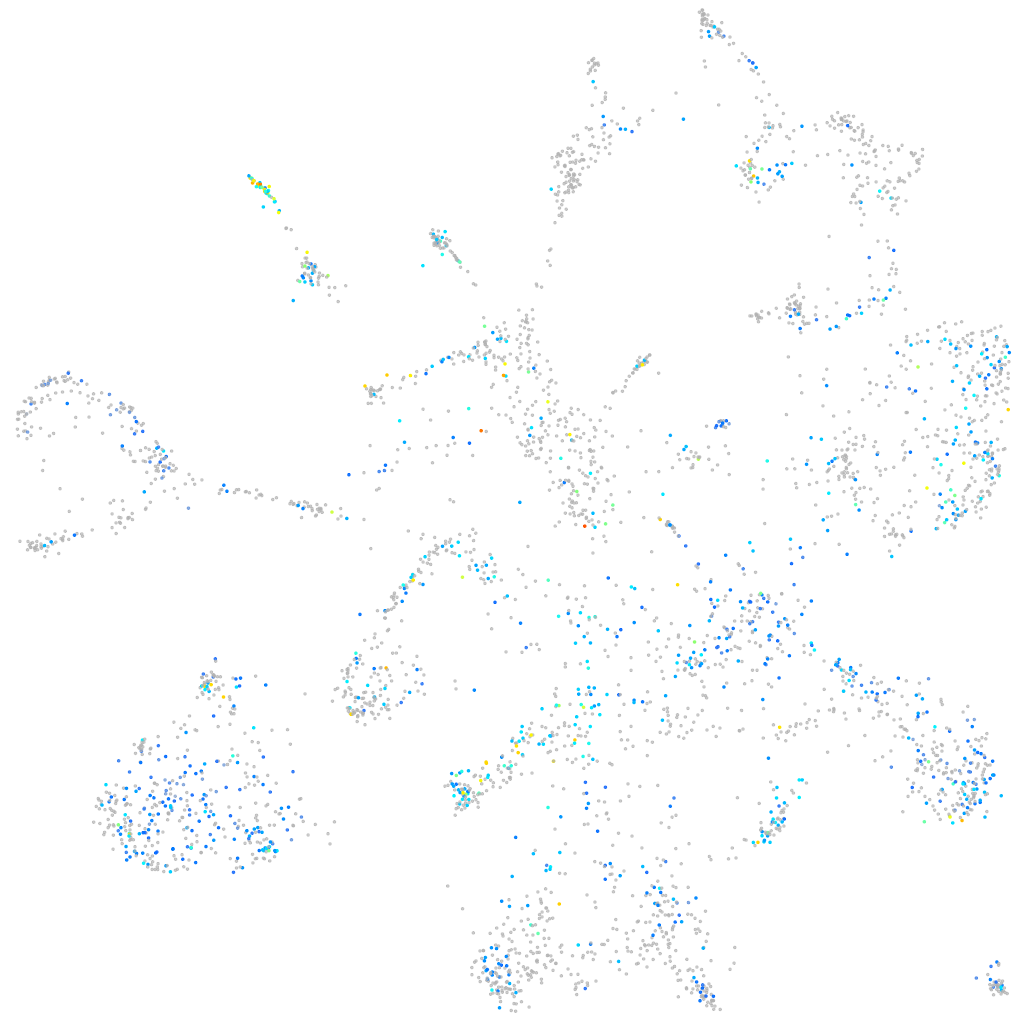
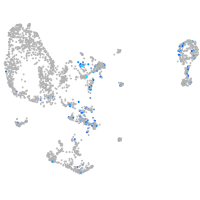
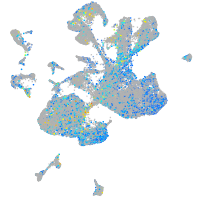
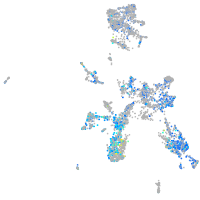
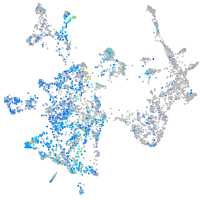
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| clec14a | 0.166 | tagln | -0.103 |
| emx2 | 0.164 | acta2 | -0.093 |
| pmp22a | 0.151 | csrp1b | -0.092 |
| mdka | 0.140 | flna | -0.085 |
| fam19a5a | 0.139 | lmod1b | -0.084 |
| lmx1bb | 0.138 | alcama | -0.084 |
| nid2a | 0.138 | rbm24a | -0.079 |
| rspo2 | 0.137 | loxa | -0.078 |
| pitx2 | 0.135 | gapdhs | -0.077 |
| mmp16a | 0.129 | pkma | -0.075 |
| si:dkey-33c9.6 | 0.126 | tpm1 | -0.075 |
| tfap2b | 0.126 | rpl38 | -0.075 |
| BX324132.3 | 0.125 | bves | -0.074 |
| lrrc17 | 0.125 | pim1 | -0.074 |
| tfap2a | 0.125 | acta1a | -0.073 |
| marcksl1a | 0.124 | cnn1b | -0.072 |
| ubc | 0.123 | phlda2 | -0.069 |
| zgc:66479 | 0.123 | slc9a3r1a | -0.069 |
| ccl38.1 | 0.123 | lpp | -0.069 |
| pdgfra | 0.123 | htra1a | -0.069 |
| fap | 0.121 | aldocb | -0.067 |
| map2 | 0.120 | bgna | -0.067 |
| csnk1a1 | 0.119 | tpm2 | -0.066 |
| hmgn2 | 0.118 | fxyd1 | -0.066 |
| hic1 | 0.117 | emid1 | -0.066 |
| msi2a | 0.117 | si:ch211-1a19.3 | -0.066 |
| inka1a | 0.117 | slc29a1b | -0.066 |
| BX248318.1 | 0.116 | ldha | -0.066 |
| foxp4 | 0.113 | pgam1a | -0.066 |
| ccdc187 | 0.112 | si:ch211-137i24.10 | -0.065 |
| si:ch211-108d22.2 | 0.112 | fhl2a | -0.065 |
| prkcab | 0.111 | tmsb4x | -0.065 |
| mt-co2 | 0.110 | glipr2l | -0.065 |
| sept2 | 0.109 | si:dkey-164f24.2 | -0.065 |
| CR383676.1 | 0.107 | gata5 | -0.064 |