ADAM metallopeptidase domain 8a
ZFIN 
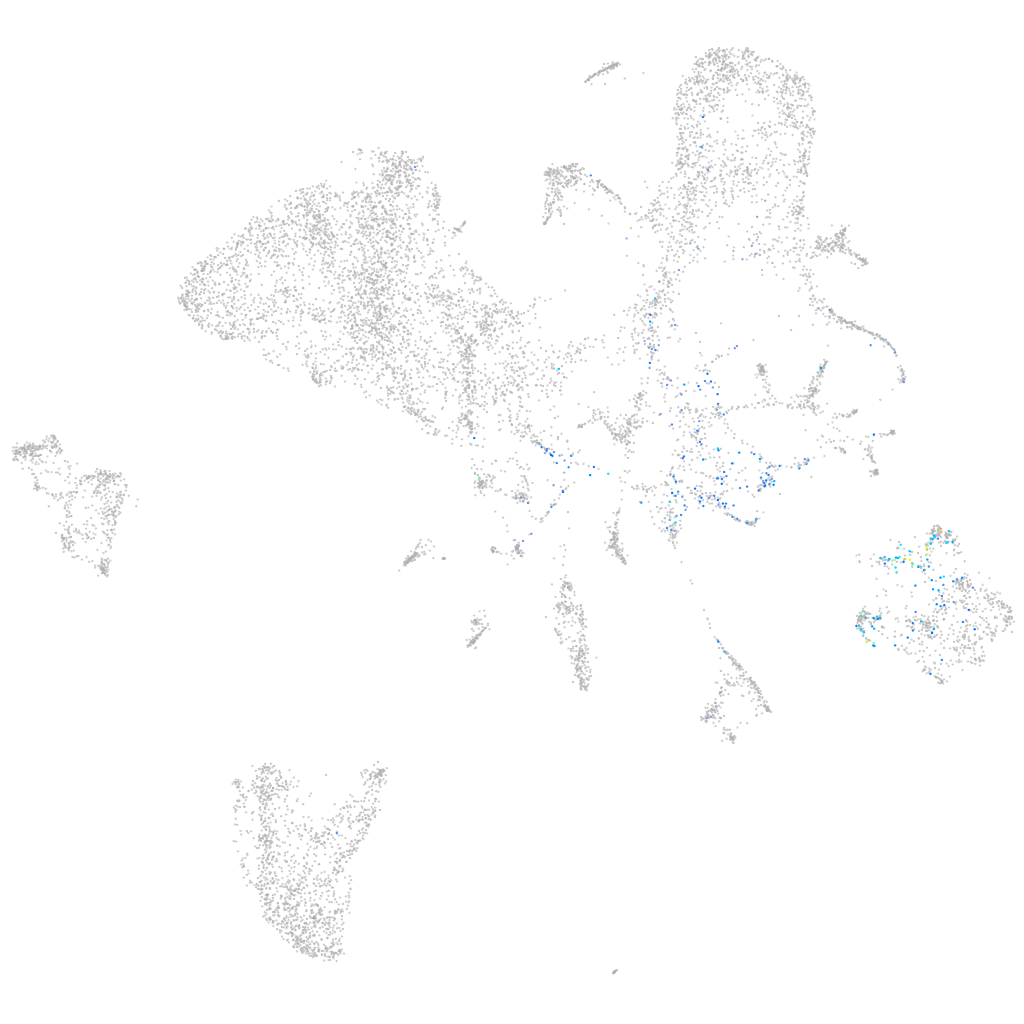
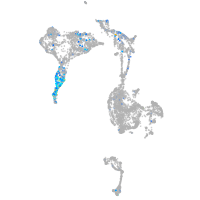
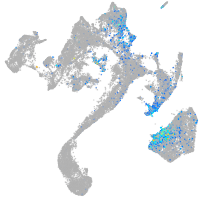
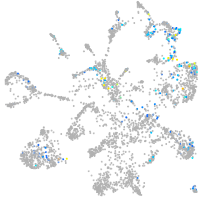
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apela | 0.273 | gapdh | -0.163 |
| nradd | 0.252 | rpl37 | -0.157 |
| pltp | 0.247 | ahcy | -0.153 |
| cdx4 | 0.246 | aldob | -0.147 |
| fgf8a | 0.230 | rps10 | -0.145 |
| cx43.4 | 0.229 | gamt | -0.144 |
| qkia | 0.224 | eno3 | -0.141 |
| hspb1 | 0.224 | sod1 | -0.140 |
| cdh6 | 0.221 | rps17 | -0.136 |
| BX001014.2 | 0.217 | eef1da | -0.134 |
| tgif1 | 0.217 | zgc:114188 | -0.134 |
| fgfrl1b | 0.215 | atp5if1b | -0.133 |
| marcksb | 0.214 | nme2b.1 | -0.131 |
| nucks1a | 0.214 | gatm | -0.129 |
| hoxc8a | 0.213 | dap | -0.128 |
| seta | 0.203 | suclg1 | -0.125 |
| hoxb7a | 0.203 | glud1b | -0.124 |
| hoxc6b | 0.202 | gpx4a | -0.124 |
| wnt5b | 0.201 | fbp1b | -0.122 |
| acin1a | 0.201 | tpi1b | -0.122 |
| si:ch73-281n10.2 | 0.200 | apoa4b.1 | -0.119 |
| mcama | 0.199 | bhmt | -0.117 |
| hmgn6 | 0.196 | nupr1b | -0.117 |
| hmgn7 | 0.194 | apoa1b | -0.115 |
| nr6a1a | 0.194 | mat1a | -0.115 |
| oc90 | 0.193 | apoc2 | -0.114 |
| si:ch211-222l21.1 | 0.193 | scp2a | -0.114 |
| hmga1a | 0.191 | mdh1aa | -0.113 |
| hnrnpaba | 0.190 | zgc:158463 | -0.113 |
| tpm4a | 0.189 | zgc:92744 | -0.112 |
| syncrip | 0.189 | gstp1 | -0.111 |
| serpinh1b | 0.188 | afp4 | -0.111 |
| NC-002333.4 | 0.187 | lgals2b | -0.110 |
| crtap | 0.185 | cx32.3 | -0.110 |
| si:ch73-1a9.3 | 0.183 | pklr | -0.110 |