zgc:158689
ZFIN 
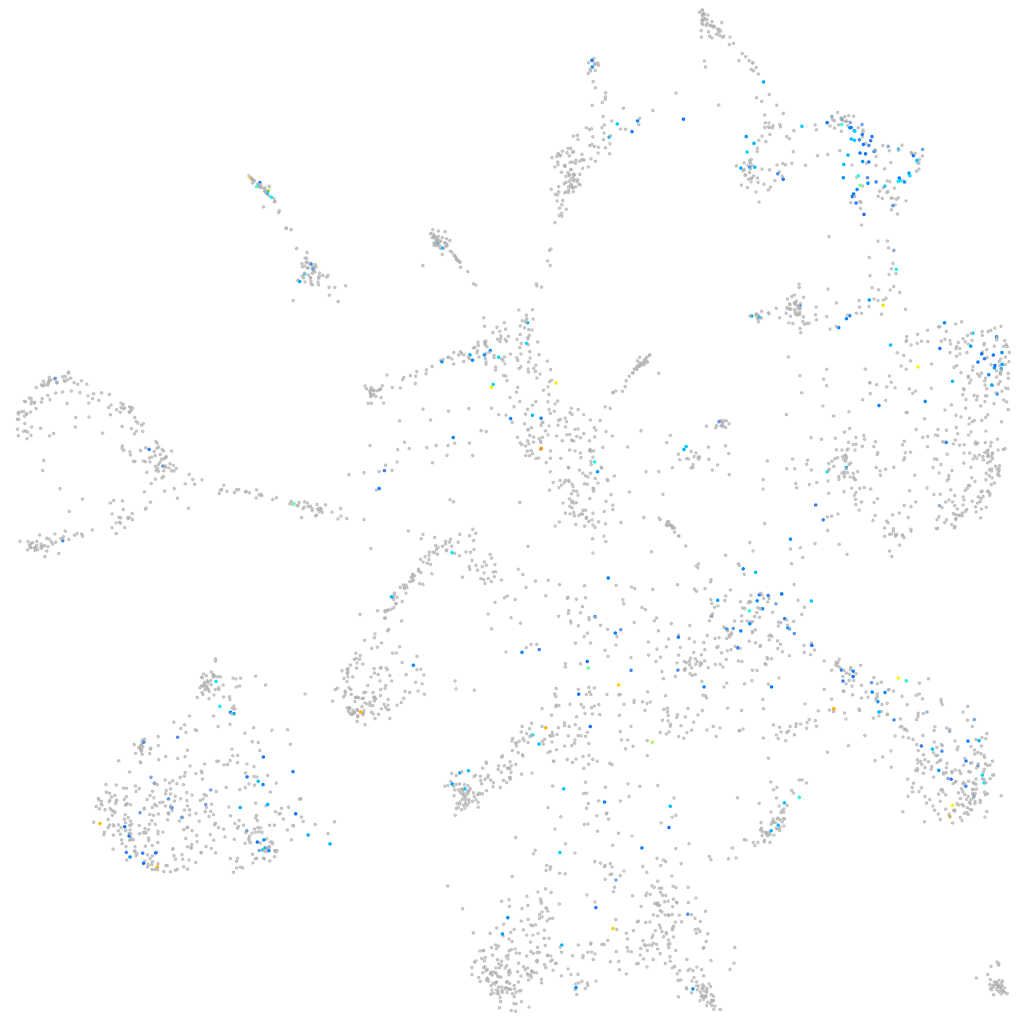
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sfrp5 | 0.201 | tpm1 | -0.125 |
| alpi.1 | 0.169 | myl9a | -0.116 |
| tnni1b | 0.155 | acta2 | -0.111 |
| CABZ01075068.1 | 0.149 | gapdhs | -0.091 |
| lin28a | 0.139 | tagln | -0.090 |
| ptger4b | 0.137 | myh11a | -0.087 |
| XLOC-001964 | 0.135 | ak1 | -0.083 |
| mdka | 0.133 | si:ch211-62a1.3 | -0.078 |
| mmel1 | 0.131 | csrp1b | -0.077 |
| tbx5a | 0.126 | gucy1a1 | -0.077 |
| XLOC-042222 | 0.125 | zgc:153704 | -0.076 |
| lmod2a | 0.124 | ptmaa | -0.076 |
| emp3b | 0.124 | fhl2a | -0.076 |
| nrip1b | 0.124 | col6a1 | -0.074 |
| BX005396.3 | 0.123 | tpm2 | -0.073 |
| CABZ01076737.1 | 0.123 | aoc2 | -0.073 |
| lbx2 | 0.123 | cald1b | -0.072 |
| thy1 | 0.121 | mylkb | -0.072 |
| alx4a | 0.116 | mylka | -0.070 |
| angpt1 | 0.116 | ckbb | -0.069 |
| vip | 0.114 | pkma | -0.068 |
| atp2a2a | 0.113 | col6a2 | -0.068 |
| gpd1a | 0.113 | cnn1b | -0.067 |
| CR848705.1 | 0.112 | fhl1a | -0.067 |
| zgc:171452 | 0.111 | pfn1 | -0.067 |
| CU896541.1 | 0.111 | eno1a | -0.067 |
| clec19a | 0.111 | XLOC-041870 | -0.066 |
| synpo2lb | 0.110 | gucy1b1 | -0.065 |
| gucy2c | 0.110 | col5a2a | -0.065 |
| zmp:0000001020 | 0.109 | myl6 | -0.065 |
| nexn | 0.108 | gpia | -0.065 |
| tbx1 | 0.107 | smtna | -0.064 |
| slc29a1b | 0.107 | aldocb | -0.064 |
| COLEC10 | 0.107 | eno3 | -0.064 |
| si:dkey-27o4.1 | 0.106 | ckba | -0.064 |