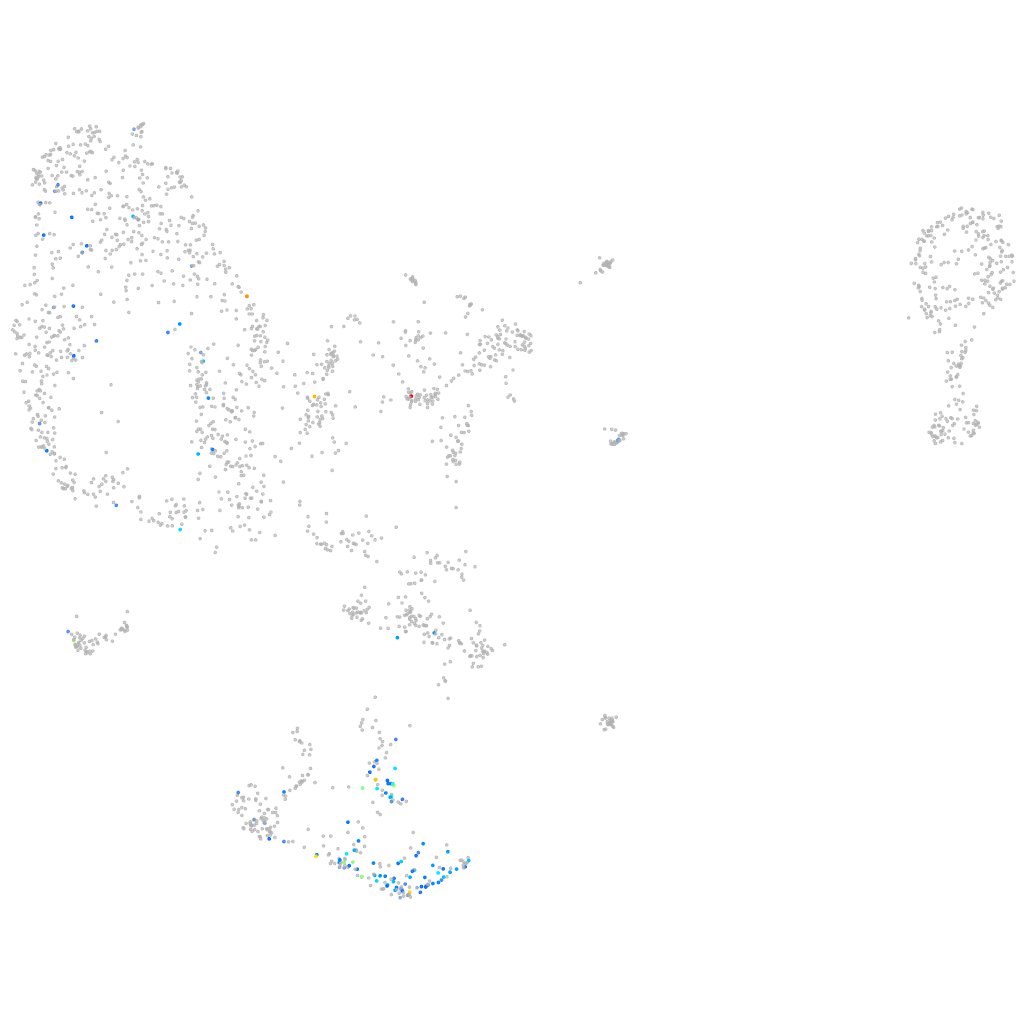
zgc:158222 [Source:ZFIN;Acc:ZDB-GENE-070112-1522]
ZFIN 
Other cell groups


all cells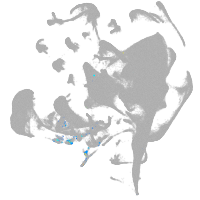 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 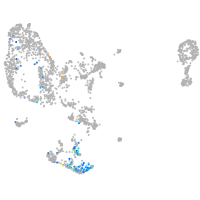 |
neural |
spinal cord / glia |
eye |
taste / olfactory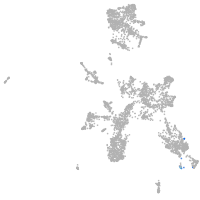 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-36i7.3 | 0.406 | acy3.2 | -0.129 |
| atp1a1a.3 | 0.392 | hnrnpabb | -0.120 |
| soul2 | 0.392 | si:ch211-288g17.3 | -0.118 |
| ca4c | 0.380 | snrpf | -0.118 |
| etf1a | 0.360 | eif5a2 | -0.117 |
| trim35-12 | 0.355 | marcksl1b | -0.115 |
| ppp1r3ab | 0.355 | pdzk1 | -0.111 |
| zgc:158423 | 0.350 | aqp8b | -0.110 |
| ppp1r1b | 0.346 | grhprb | -0.109 |
| si:ch73-359m17.9 | 0.345 | slc9a3r1a | -0.108 |
| glsb | 0.337 | si:dkey-194e6.1 | -0.105 |
| bsnd | 0.331 | pfdn4 | -0.104 |
| ckmt1 | 0.328 | hspb1 | -0.104 |
| mal2 | 0.326 | gnpda1 | -0.103 |
| agr2 | 0.321 | snu13b | -0.102 |
| slc9a3.2 | 0.320 | dab2 | -0.100 |
| CABZ01044023.1 | 0.319 | h2afvb | -0.100 |
| erbb3b | 0.318 | si:ch73-1a9.3 | -0.100 |
| gdpd3a | 0.318 | si:ch73-281n10.2 | -0.100 |
| tmem266 | 0.317 | msna | -0.100 |
| rnf183 | 0.312 | fxyd6l | -0.100 |
| trim101 | 0.307 | ptges3b | -0.100 |
| tbx2b | 0.305 | snrpd2 | -0.099 |
| phlda2 | 0.299 | fbl | -0.099 |
| mctp2b | 0.297 | fbp1b | -0.099 |
| rhcgb | 0.294 | cldn15a | -0.099 |
| me3 | 0.290 | anp32e | -0.098 |
| clcnk | 0.289 | marcksb | -0.097 |
| alox5a | 0.288 | lrp2a | -0.096 |
| slc7a8a | 0.287 | npm1a | -0.096 |
| tmem72 | 0.287 | smarce1 | -0.096 |
| cd63 | 0.286 | hmgb2b | -0.096 |
| slc12a3 | 0.281 | slc4a4a | -0.096 |
| sorl1 | 0.280 | slc26a1 | -0.096 |
| cldnh | 0.277 | syncrip | -0.096 |