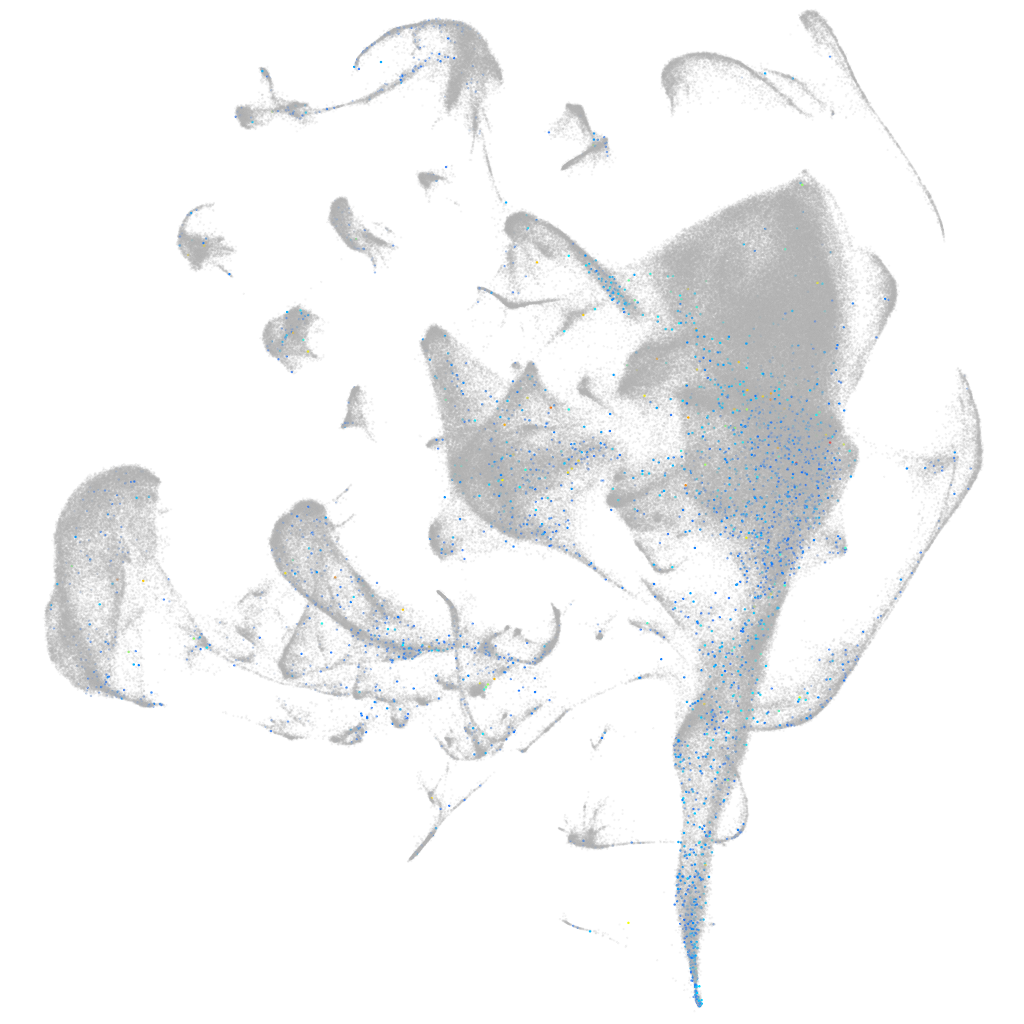zgc:103564
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccna2 | 0.094 | rtn1a | -0.034 |
| mki67 | 0.088 | gng3 | -0.033 |
| chaf1a | 0.082 | elavl3 | -0.032 |
| ccnb1 | 0.081 | pvalb2 | -0.031 |
| pcna | 0.081 | stmn1b | -0.031 |
| tpx2 | 0.081 | atp6v0cb | -0.030 |
| banf1 | 0.080 | pvalb1 | -0.030 |
| rrm1 | 0.080 | sncb | -0.030 |
| cks1b | 0.079 | actc1b | -0.029 |
| stmn1a | 0.079 | calm1a | -0.029 |
| mad2l1 | 0.078 | gpm6aa | -0.028 |
| nasp | 0.077 | rnasekb | -0.028 |
| dek | 0.076 | tmsb | -0.028 |
| fbxo5 | 0.076 | vamp2 | -0.028 |
| smc4 | 0.076 | ccni | -0.027 |
| smc2 | 0.075 | fez1 | -0.027 |
| anp32b | 0.074 | gpm6ab | -0.027 |
| dut | 0.074 | rtn1b | -0.027 |
| lbr | 0.074 | ywhag2 | -0.027 |
| nanog | 0.074 | stxbp1a | -0.027 |
| ncapg | 0.074 | elavl4 | -0.026 |
| rpa3 | 0.074 | mllt11 | -0.026 |
| ccna1 | 0.072 | si:dkeyp-75h12.5 | -0.026 |
| h1m | 0.072 | stx1b | -0.026 |
| aurkb | 0.071 | atp6v1e1b | -0.025 |
| btg4 | 0.071 | ckbb | -0.025 |
| cdca8 | 0.071 | dpysl3 | -0.025 |
| cenpw | 0.071 | stmn2a | -0.025 |
| zgc:113424 | 0.071 | tob1b | -0.025 |
| ccnb2 | 0.070 | zgc:65894 | -0.025 |
| lig1 | 0.070 | gap43 | -0.024 |
| pttg1 | 0.070 | gng2 | -0.024 |
| zgc:173742 | 0.070 | kdm6bb | -0.024 |
| cdk1 | 0.069 | nova2 | -0.024 |
| dhfr | 0.069 | snap25a | -0.024 |