zgc:103564
ZFIN 
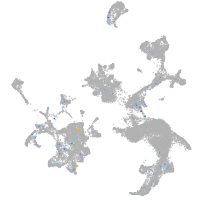
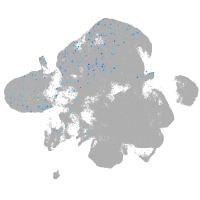
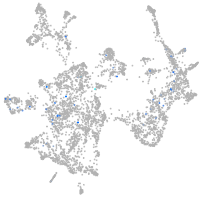
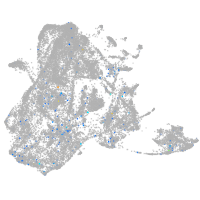
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR788316.4 | 0.113 | rtn1a | -0.037 |
| CR848665.1 | 0.097 | si:dkeyp-75h12.5 | -0.033 |
| BX649490.1 | 0.090 | marcksl1b | -0.032 |
| BX324216.4 | 0.087 | dpysl3 | -0.032 |
| CR936408.2 | 0.082 | gng3 | -0.031 |
| XLOC-028457 | 0.081 | elavl3 | -0.031 |
| LOC103911588 | 0.080 | ckbb | -0.031 |
| mki67 | 0.079 | sncb | -0.031 |
| LOC108179963 | 0.078 | gpm6aa | -0.030 |
| BX571701.1 | 0.078 | pvalb1 | -0.030 |
| CT027805.1 | 0.078 | mllt11 | -0.030 |
| ssuh2.2 | 0.077 | gpm6ab | -0.030 |
| LOC570903 | 0.076 | tmsb | -0.029 |
| ccna2 | 0.075 | gapdhs | -0.029 |
| cks1b | 0.074 | stmn1b | -0.029 |
| CABZ01051600.1 | 0.072 | atp6v0cb | -0.029 |
| rrm2 | 0.072 | nsg2 | -0.029 |
| pcna | 0.070 | fez1 | -0.028 |
| LOC110439597 | 0.070 | stxbp1a | -0.028 |
| LOC110439870 | 0.069 | vamp2 | -0.028 |
| esco2 | 0.069 | myt1b | -0.028 |
| rrm1 | 0.068 | elavl4 | -0.027 |
| XLOC-040355 | 0.068 | stx1b | -0.027 |
| dut | 0.067 | calm1a | -0.027 |
| tpx2 | 0.066 | atp6v1e1b | -0.027 |
| mad2l1 | 0.066 | pvalb2 | -0.027 |
| lbr | 0.065 | hbae3 | -0.027 |
| aurkb | 0.064 | rnasekb | -0.026 |
| mcm7 | 0.064 | cotl1 | -0.026 |
| top2a | 0.064 | si:ch211-260e23.9 | -0.026 |
| spc24 | 0.064 | cplx2 | -0.026 |
| cdk1 | 0.064 | ywhag2 | -0.025 |
| armc1l | 0.064 | ppdpfb | -0.025 |
| kifc1 | 0.063 | gabarapl2 | -0.025 |
| nutf2l | 0.063 | cadm4 | -0.025 |