"zinc finger protein, FOG family member 1"
ZFIN 
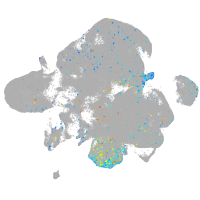
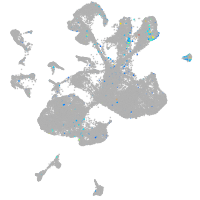
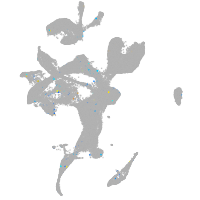
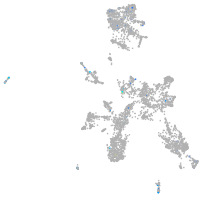
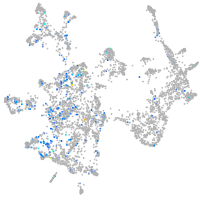
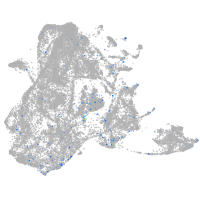
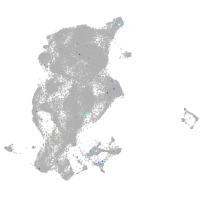
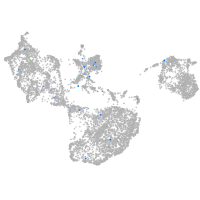
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gata5 | 0.161 | fibina | -0.040 |
| gata6 | 0.113 | col2a1b | -0.038 |
| tnni1b | 0.111 | fgfbp2b | -0.037 |
| podxl | 0.110 | col9a2 | -0.037 |
| tmem88b | 0.107 | col9a3 | -0.037 |
| fbn2a.1 | 0.102 | col9a1a | -0.036 |
| mmel1 | 0.100 | bhmt | -0.035 |
| BX469925.3 | 0.098 | vwde | -0.035 |
| XLOC-028299 | 0.098 | eef1da | -0.035 |
| cracr2b | 0.096 | col11a2 | -0.035 |
| tmem98 | 0.094 | col2a1a | -0.034 |
| tdgf1 | 0.094 | nme2b.1 | -0.033 |
| ftr82 | 0.094 | cnmd | -0.033 |
| tnnt2a | 0.088 | ecrg4a | -0.031 |
| crb2a | 0.087 | col11a1a | -0.031 |
| meis3 | 0.086 | tgfbi | -0.030 |
| XLOC-015291 | 0.085 | tnn | -0.030 |
| jam2b | 0.084 | CU929237.1 | -0.030 |
| si:dkey-13e3.1 | 0.083 | rps29 | -0.030 |
| alcamb | 0.081 | serpinf1 | -0.029 |
| hspb1 | 0.081 | matn1 | -0.029 |
| rbpms2b | 0.080 | VIT | -0.029 |
| myl7 | 0.079 | sparc | -0.029 |
| synpo2lb | 0.079 | epyc | -0.029 |
| nkx2.5 | 0.079 | fkbp11 | -0.028 |
| LOC569612 | 0.078 | rpl37 | -0.028 |
| gdf3 | 0.077 | cspg4 | -0.028 |
| XLOC-015407 | 0.077 | sox9a | -0.028 |
| CABZ01075068.1 | 0.076 | rpl38 | -0.027 |
| tmem108 | 0.074 | mia | -0.027 |
| foxh1 | 0.074 | XLOC-035413 | -0.027 |
| rbpms2a | 0.073 | rpl29 | -0.027 |
| tal2 | 0.072 | dlx3b | -0.027 |
| rgs14a | 0.070 | copz2 | -0.026 |
| nkx2.7 | 0.070 | CYTL1 | -0.026 |